|
chr6_+_36098261
|
0.820
|
NM_002754
|
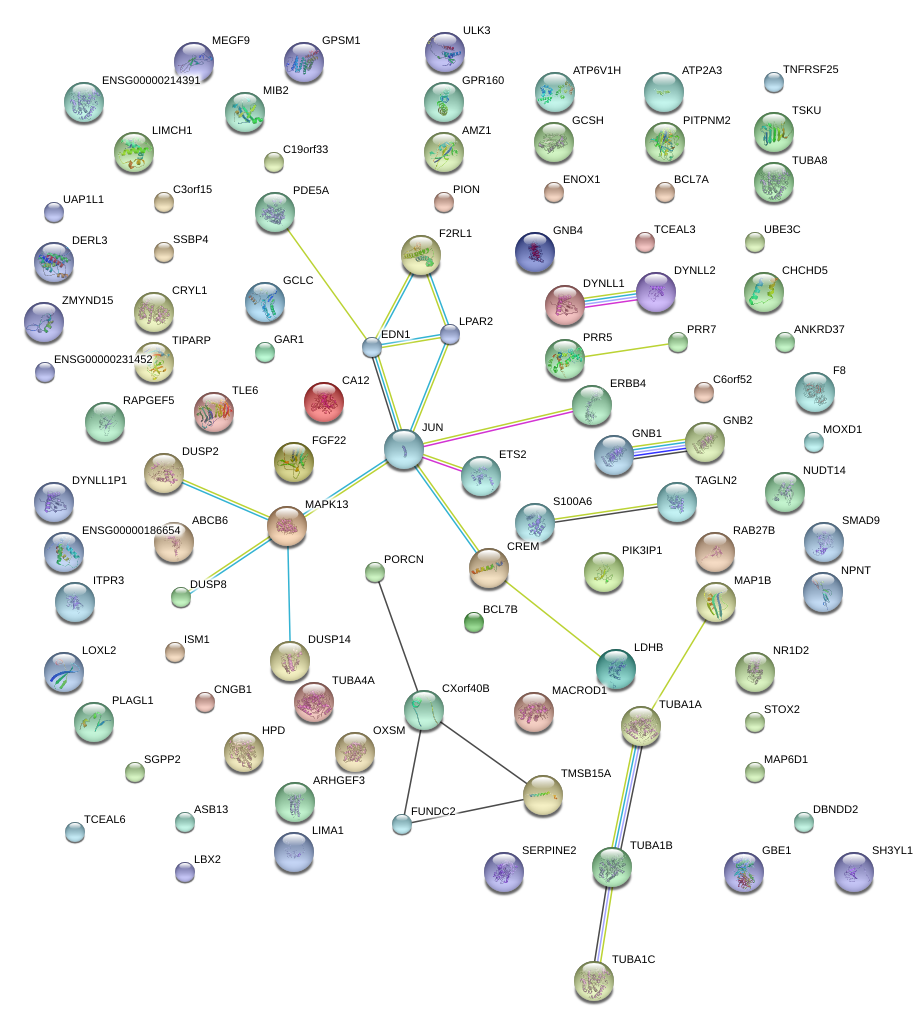
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr5_+_176873795
|
0.565
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chrX_-_148622357
|
0.565
|
|
LOC100131434
|
uncharacterized LOC100131434
|
|
chr4_+_186317964
|
0.523
|
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr7_+_2719155
|
0.445
|
NM_133463
|
AMZ1
|
archaelysin family metallopeptidase 1
|
|
chr6_+_12290528
|
0.423
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr5_+_76114881
|
0.392
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr2_-_110970103
|
0.385
|
|
LINC00116
|
long intergenic non-protein coding RNA 116
|
|
chr11_-_63933493
|
0.348
|
NM_014067
|
MACROD1
|
MACRO domain containing 1
|
|
chr20_-_656443
|
0.343
|
|
|
|
|
chr20_+_13202417
|
0.339
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr6_-_10694837
|
0.330
|
NM_001145020
|
C6orf52
|
chromosome 6 open reading frame 52
|
|
chr6_+_33589045
|
0.330
|
NM_002224
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3
|
|
chr1_-_1551041
|
0.296
|
|
MIB2
|
mindbomb homolog 2 (Drosophila)
|
|
chr12_-_21810720
|
0.280
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_49582800
|
0.279
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr17_+_17398334
|
0.264
|
|
|
|
|
chr5_+_76114832
|
0.263
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chrX_-_154255214
|
0.262
|
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr19_+_38794800
|
0.251
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr10_-_98592186
|
0.246
|
|
|
|
|
chr12_-_122326516
|
0.237
|
NM_001171993
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr2_-_200715895
|
0.235
|
|
FONG
|
uncharacterized LOC348751
|
|
chr8_-_54755836
|
0.235
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr4_+_186317837
|
0.231
|
NM_181726
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr15_-_75135524
|
0.227
|
NM_001099436
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr13_-_37494370
|
0.225
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr1_-_153508463
|
0.221
|
|
S100A6
|
S100 calcium binding protein A6
|
|
chr7_+_27135672
|
0.219
|
|
HOTAIRM1
|
HOXA transcript antisense RNA, myeloid-specific 1 (non-protein coding)
|
|
chr12_-_50677238
|
0.218
|
NM_001113546
NM_016357
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr19_+_2977480
|
0.216
|
NM_001143986
NM_024760
|
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila)
|
|
chr12_-_21810754
|
0.209
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21810769
|
0.209
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr13_-_44361032
|
0.208
|
NM_001242863
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr2_-_264037
|
0.206
|
NM_001159597
NM_015677
|
SH3YL1
|
SH3 domain containing, Ysc84-like 1 (S. cerevisiae)
|
|
chr11_-_63933204
|
0.204
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr19_-_19738973
|
0.204
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr7_-_72972021
|
0.203
|
NM_001197244
NM_001707
|
BCL7B
|
B-cell CLL/lymphoma 7B
|
|
chr6_-_28186663
|
0.202
|
|
TOB2P1
|
transducer of ERBB2, 2 pseudogene 1
|
|
chr6_-_53409795
|
0.201
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr20_+_44035203
|
0.199
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr7_-_22396511
|
0.199
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr17_-_3867585
|
0.199
|
NM_005173
NM_174953
NM_174954
NM_174955
NM_174956
NM_174957
NM_174958
|
ATP2A3
|
ATPase, Ca++ transporting, ubiquitous
|
|
chr3_-_179169180
|
0.196
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr15_-_75135480
|
0.194
|
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr2_-_96810775
|
0.192
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr3_-_57113292
|
0.191
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr7_-_77045706
|
0.191
|
NM_017439
|
PION
|
pigeon homolog (Drosophila)
|
|
chr6_-_53409438
|
0.187
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr8_+_128806720
|
0.187
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr1_-_144520918
|
0.186
|
|
LOC728855
|
uncharacterized LOC728855
|
|
chr4_+_184826427
|
0.186
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr3_-_81810802
|
0.184
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr6_-_53409507
|
0.184
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr6_-_53409834
|
0.182
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr1_-_6526150
|
0.178
|
NM_001039664
NM_003790
NM_148965
NM_148966
NM_148967
NM_148970
|
TNFRSF25
|
tumor necrosis factor receptor superfamily, member 25
|
|
chr11_+_76494132
|
0.175
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr21_+_40177754
|
0.175
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr2_-_74730342
|
0.175
|
NM_001009812
|
LBX2
|
ladybird homeobox 2
|
|
chr4_+_41362565
|
0.174
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr8_-_54755821
|
0.173
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr8_-_23261632
|
0.173
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr19_+_18530211
|
0.172
|
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr22_-_24181173
|
0.170
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chr3_+_169755733
|
0.170
|
NM_014373
|
GPR160
|
G protein-coupled receptor 160
|
|
chr2_-_220083394
|
0.170
|
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr13_-_21099914
|
0.169
|
NM_015974
|
CRYL1
|
crystallin, lambda 1
|
|
chr11_-_1593149
|
0.169
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr4_+_41362750
|
0.168
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr8_-_54755546
|
0.168
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr7_+_156931609
|
0.167
|
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr12_-_123635284
|
0.165
|
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr19_+_639878
|
0.165
|
NM_020637
|
FGF22
|
fibroblast growth factor 22
|
|
chr3_-_183543295
|
0.164
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr21_+_40177847
|
0.163
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr2_-_70314065
|
0.163
|
|
PCBP1-AS1
|
PCBP1 antisense RNA 1 (non-protein coding)
|
|
chr4_+_106816593
|
0.162
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr2_-_224903933
|
0.161
|
NM_001136528
NM_006216
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr17_+_4643318
|
0.160
|
NM_001136046
NM_032265
|
ZMYND15
|
zinc finger, MYND-type containing 15
|
|
chr6_-_144329254
|
0.160
|
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chrX_+_148622518
|
0.160
|
NM_001171907
NM_001171908
NM_178124
|
CXorf40A
|
chromosome X open reading frame 40A
|
|
chr4_-_120548329
|
0.159
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr10_+_35416384
|
0.157
|
NM_181571
NM_183060
|
CREM
|
cAMP responsive element modulator
|
|
chr2_+_113342171
|
0.156
|
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chr4_+_110736911
|
0.156
|
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr3_+_23987514
|
0.155
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_-_179169330
|
0.155
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr7_+_156803599
|
0.154
|
|
LOC645249
|
uncharacterized LOC645249
|
|
chr9_+_139972195
|
0.154
|
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr16_-_81129882
|
0.152
|
NM_004483
|
GCSH
|
glycine cleavage system protein H (aminomethyl carrier)
|
|
chrX_+_154255063
|
0.151
|
NM_023934
|
FUNDC2
|
FUN14 domain containing 2
|
|
chr15_-_75135479
|
0.151
|
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chrX_+_102862833
|
0.149
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr9_-_123476578
|
0.148
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr10_-_28821459
|
0.147
|
|
LOC220906
|
uncharacterized LOC220906
|
|
chr15_-_75135429
|
0.147
|
|
ULK3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr3_+_52273247
|
0.146
|
|
|
|
|
chr9_+_139971938
|
0.146
|
NM_207309
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr15_-_63673918
|
0.146
|
|
CA12
|
carbonic anhydrase XII
|
|
chr3_+_25831548
|
0.145
|
NM_001145391
NM_017897
|
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial
|
|
chr3_+_169755782
|
0.145
|
|
GPR160
|
G protein-coupled receptor 160
|
|
chr3_-_161089747
|
0.145
|
NM_001040100
|
SPTSSB
|
serine palmitoyltransferase, small subunit B
|
|
chr1_-_59249687
|
0.144
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chr1_-_159895258
|
0.143
|
|
TAGLN2
|
transgelin 2
|
|
chr10_-_5708542
|
0.141
|
NM_024701
|
ASB13
|
ankyrin repeat and SOCS box containing 13
|
|
chrX_-_101771641
|
0.141
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chrX_+_48367346
|
0.141
|
NM_022825
NM_203473
|
PORCN
|
porcupine homolog (Drosophila)
|
|
chr3_+_119421865
|
0.141
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr2_+_223289285
|
0.140
|
NM_152386
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr1_-_175161832
|
0.140
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr2_-_213403280
|
0.139
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr3_+_159943628
|
0.138
|
|
C3orf80
|
chromosome 3 open reading frame 80
|
|
chr22_-_31686707
|
0.138
|
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr4_+_186317892
|
0.137
|
|
ANKRD37
|
ankyrin repeat domain 37
|
|
chr3_-_81810914
|
0.137
|
NM_000158
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr6_-_132722564
|
0.135
|
NM_015529
|
MOXD1
|
monooxygenase, DBH-like 1
|
|
chr2_-_220083658
|
0.135
|
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr16_-_3109335
|
0.134
|
|
LOC100507419
|
uncharacterized LOC100507419
|
|
chr1_-_159895279
|
0.134
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chr9_+_139247508
|
0.133
|
NM_001145639
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr14_-_105647520
|
0.133
|
NM_177533
|
NUDT14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14
|
|
chr22_+_45148437
|
0.132
|
NM_001017526
NM_001198726
NM_181335
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr18_+_52495618
|
0.132
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr17_+_35849997
|
0.132
|
|
DUSP14
|
dual specificity phosphatase 14
|
|
chr7_-_72971868
|
0.131
|
|
BCL7B
|
B-cell CLL/lymphoma 7B
|
|
chr3_+_156392369
|
0.131
|
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr1_-_59248287
|
0.131
|
|
JUN
|
jun proto-oncogene
|
|
chr12_+_120933911
|
0.130
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr5_+_71403040
|
0.130
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr12_+_120933916
|
0.130
|
|
DYNLL1
|
dynein, light chain, LC8-type 1
|
|
chr19_+_56989549
|
0.130
|
|
LOC100128252
|
uncharacterized LOC100128252
|
|
chr11_-_74204708
|
0.129
|
NM_001144869
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative)
|
|
chr12_+_3068989
|
0.129
|
|
TEAD4
|
TEA domain family member 4
|
|
chr18_+_9914000
|
0.128
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr12_-_21810733
|
0.128
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr9_-_72287097
|
0.128
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr20_+_48599511
|
0.128
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr19_+_39897457
|
0.127
|
NM_003407
|
ZFP36
|
zinc finger protein 36, C3H type, homolog (mouse)
|
|
chr17_-_63052884
|
0.127
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr7_+_17338182
|
0.126
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr3_-_133614421
|
0.126
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr19_+_11039419
|
0.126
|
NM_138358
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr19_+_10654570
|
0.126
|
|
ATG4D
|
ATG4 autophagy related 4 homolog D (S. cerevisiae)
|
|
chr8_-_23261626
|
0.125
|
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr17_-_63052744
|
0.125
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr12_-_117175634
|
0.125
|
NM_024738
|
C12orf49
|
chromosome 12 open reading frame 49
|
|
chr19_+_1026582
|
0.125
|
|
CNN2
|
calponin 2
|
|
chr15_-_42264749
|
0.124
|
NM_139265
|
EHD4
|
EH-domain containing 4
|
|
chr1_+_861083
|
0.123
|
NM_152486
|
SAMD11
|
sterile alpha motif domain containing 11
|
|
chr14_+_55034633
|
0.123
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr17_-_66287256
|
0.122
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr6_-_144385647
|
0.122
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr19_+_18391252
|
0.122
|
|
|
|
|
chr12_-_21810779
|
0.121
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chr16_+_68771214
|
0.120
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr3_-_81810816
|
0.120
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr11_+_1331048
|
0.120
|
|
LOC255512
|
uncharacterized LOC255512
|
|
chr16_-_54962707
|
0.120
|
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr16_+_68771192
|
0.119
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr2_-_220083699
|
0.119
|
NM_005689
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr8_+_22298767
|
0.119
|
|
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme
|
|
chr8_+_31497267
|
0.118
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr17_-_79479831
|
0.118
|
NM_001199954
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr14_+_55034329
|
0.118
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr12_-_117175844
|
0.118
|
|
C12orf49
|
chromosome 12 open reading frame 49
|
|
chrX_-_69509714
|
0.117
|
NM_016484
|
PDZD11
|
PDZ domain containing 11
|
|
chr11_+_637268
|
0.117
|
NM_000797
|
DRD4
|
dopamine receptor D4
|
|
chr17_-_15164043
|
0.116
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr17_+_64960754
|
0.116
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr1_-_8939150
|
0.115
|
NM_001428
|
ENO1
|
enolase 1, (alpha)
|
|
chr19_+_1026608
|
0.115
|
|
CNN2
|
calponin 2
|
|
chr17_-_79479780
|
0.115
|
|
ACTG1
|
actin, gamma 1
|
|
chr17_-_74497406
|
0.114
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr21_+_42688660
|
0.113
|
NM_058186
NM_206964
|
FAM3B
|
family with sequence similarity 3, member B
|
|
chr19_+_36980524
|
0.113
|
|
LOC728752
|
uncharacterized LOC728752
|
|
chr9_+_117373622
|
0.112
|
NM_153045
|
C9orf91
|
chromosome 9 open reading frame 91
|
|
chr17_-_39684499
|
0.112
|
|
KRT19
|
keratin 19
|
|
chr1_-_1710228
|
0.112
|
NM_001198993
|
NADK
|
NAD kinase
|
|
chr10_+_81070578
|
0.112
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr17_-_63052816
|
0.111
|
|
|
|
|
chr22_+_25960885
|
0.110
|
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr9_-_110251754
|
0.110
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr15_-_63674074
|
0.109
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr12_-_53473171
|
0.109
|
NM_032840
|
SPRYD3
|
SPRY domain containing 3
|
|
chr7_+_56032065
|
0.109
|
NM_001202469
NM_001483
|
GBAS
|
glioblastoma amplified sequence
|
|
chr9_-_97401588
|
0.108
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr19_-_18548788
|
0.108
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr17_-_4890920
|
0.108
|
NM_001171166
NM_015099
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr10_+_28821724
|
0.107
|
|
|
|
|
chr2_+_47596447
|
0.107
|
|
EPCAM
|
epithelial cell adhesion molecule
|
|
chr3_-_81810618
|
0.107
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr15_-_42264687
|
0.107
|
|
EHD4
|
EH-domain containing 4
|
|
chr12_+_107349673
|
0.106
|
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr6_-_144329342
|
0.106
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chr17_-_4890664
|
0.104
|
NM_001171167
NM_001171168
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr12_-_42631982
|
0.103
|
NM_001190977
|
YAF2
|
YY1 associated factor 2
|
|
chr18_-_21017816
|
0.103
|
NM_032933
|
TMEM241
|
transmembrane protein 241
|
|
chr17_-_63052707
|
0.103
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr20_+_4129466
|
0.102
|
|
SMOX
|
spermine oxidase
|
|
chr16_+_68771236
|
0.102
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|