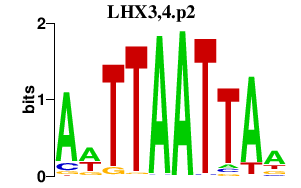
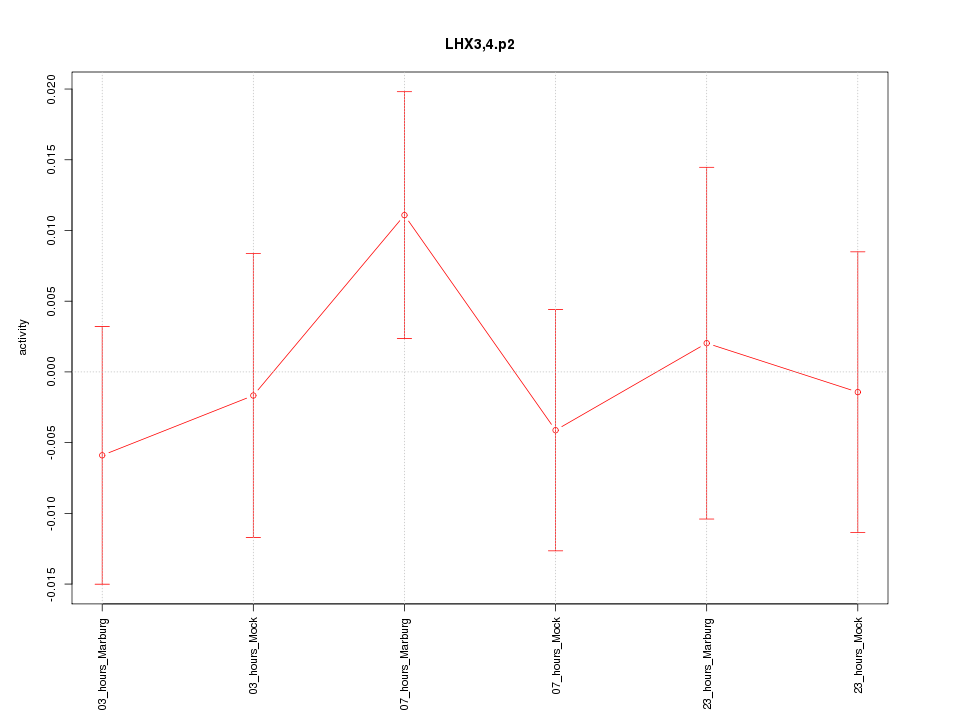
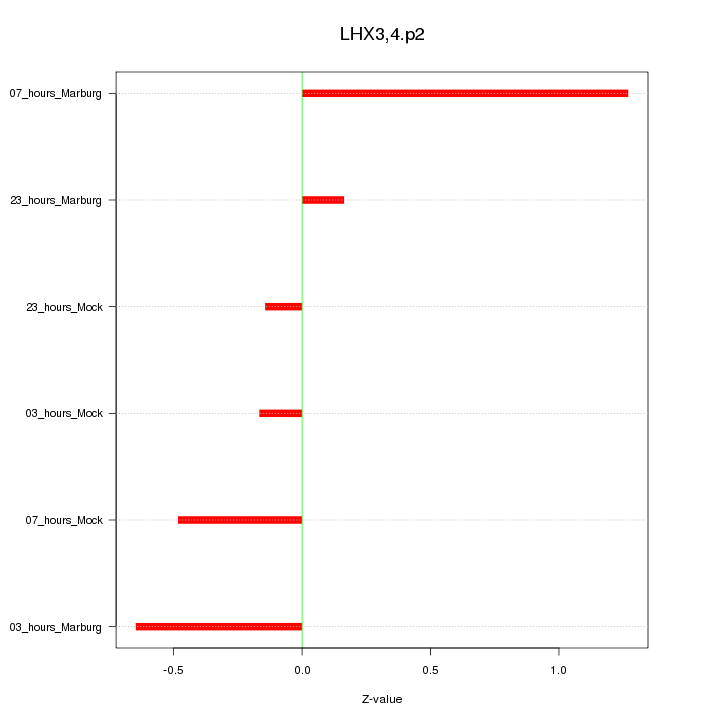
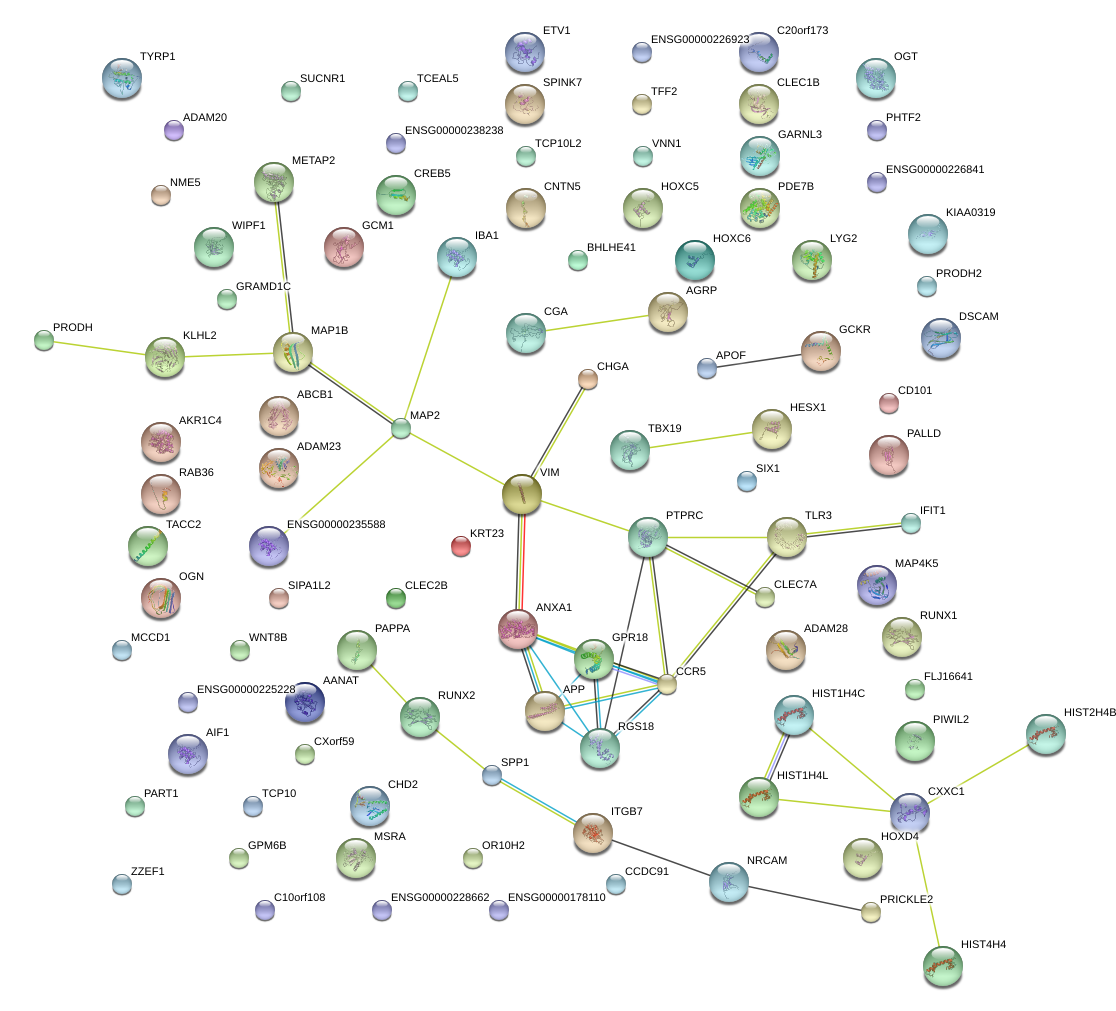
|
chr12_+_109551041
|
0.395
|
|
|
|
|
chr3_-_57234279
|
0.379
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr1_+_43253660
|
0.376
|
NM_001242750
|
LOC100129924
|
uncharacterized LOC100129924
|
|
chr9_+_12693385
|
0.372
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr3_+_151591428
|
0.351
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr5_+_147691988
|
0.318
|
NM_032566
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative)
|
|
chr8_+_32785485
|
0.301
|
|
|
|
|
chr1_+_168250277
|
0.301
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr12_-_10021932
|
0.282
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr12_-_10282680
|
0.281
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr10_+_5238756
|
0.238
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chrM_+_10123
|
0.232
|
|
|
|
|
chr6_-_87804773
|
0.224
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr2_-_99871569
|
0.209
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr10_+_695887
|
0.206
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr2_+_27719702
|
0.200
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr6_+_31496738
|
0.198
|
NM_001011700
|
MCCD1
|
mitochondrial coiled-coil domain 1
|
|
chr17_+_74463629
|
0.196
|
NM_001088
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr5_-_137475076
|
0.194
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr13_+_24463006
|
0.191
|
NM_001014442
|
C1QTNF9B-AS1
|
C1QTNF9B antisense RNA 1 (non-protein coding)
|
|
chr4_+_88896838
|
0.190
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_+_88896868
|
0.188
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr12_-_26277807
|
0.187
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr16_-_67517715
|
0.186
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chr6_-_167797917
|
0.178
|
NM_004610
|
TCP10
|
t-complex 10 homolog (mouse)
|
|
chr20_-_34117447
|
0.166
|
NM_001145350
|
C20orf173
|
chromosome 20 open reading frame 173
|
|
chr7_-_107880492
|
0.165
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr14_-_71001731
|
0.162
|
NM_003814
|
ADAM20
|
ADAM metallopeptidase domain 20
|
|
chr5_+_59784004
|
0.162
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chrX_-_102531673
|
0.157
|
NM_001012979
|
TCEAL5
|
transcription elongation factor A (SII)-like 5
|
|
chr4_+_88896899
|
0.155
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr6_+_26104103
|
0.149
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|
|
chr2_+_210444364
|
0.142
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr9_-_95166827
|
0.140
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr19_-_36304200
|
0.138
|
NM_021232
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr11_-_71524890
|
0.124
|
|
LOC285407
|
asparagine-linked glycosylation 1 homolog pseudogene
|
|
chr3_+_82035288
|
0.123
|
|
|
|
|
chr6_-_53013581
|
0.123
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chr8_+_22210311
|
0.115
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr9_+_75766777
|
0.112
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr4_+_88896801
|
0.111
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr12_-_53600999
|
0.111
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr7_+_28452143
|
0.110
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_+_46411632
|
0.106
|
NM_000579
NM_001100168
|
CCR5
|
chemokine (C-C motif) receptor 5
|
|
chr14_-_61115906
|
0.106
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr6_-_133035180
|
0.103
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr3_+_73110809
|
0.103
|
NM_018029
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2
|
|
chr19_-_36303724
|
0.102
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr13_-_99910548
|
0.100
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr12_+_54410893
|
0.099
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr4_+_186990308
|
0.098
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr1_+_117544371
|
0.098
|
NM_004258
|
CD101
|
CD101 molecule
|
|
chr7_-_14025826
|
0.096
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr2_+_177016112
|
0.096
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr1_+_192127591
|
0.093
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chrX_+_70765479
|
0.093
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chrX_-_13835187
|
0.092
|
|
GPM6B
|
glycoprotein M6B
|
|
chr2_-_175462428
|
0.092
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr4_+_166131170
|
0.092
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr1_+_97260515
|
0.091
|
|
|
|
|
chr7_-_87342569
|
0.090
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr1_+_198608216
|
0.087
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr6_-_24583454
|
0.086
|
NM_001252328
|
KIAA0319
|
KIAA0319
|
|
chr10_+_102222797
|
0.078
|
NM_003393
|
WNT8B
|
wingless-type MMTV integration site family, member 8B
|
|
chr3_+_113616317
|
0.077
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr19_+_15838833
|
0.077
|
NM_013939
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2
|
|
chr4_+_169418167
|
0.076
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr17_-_39092714
|
0.076
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr11_+_99426872
|
0.075
|
NM_175566
|
CNTN5
|
contactin 5
|
|
chr21_-_27372461
|
0.075
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr3_-_64211111
|
0.073
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr2_+_177016337
|
0.072
|
|
|
|
|
chr3_+_156544075
|
0.072
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr9_+_118916069
|
0.071
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr5_+_71502700
|
0.071
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr7_+_77469446
|
0.070
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr6_+_31583766
|
0.069
|
NM_032955
|
AIF1
|
allograft inflammatory factor 1
|
|
chr6_+_136172802
|
0.068
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr1_-_232651227
|
0.067
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr21_-_43771069
|
0.066
|
NM_005423
|
TFF2
|
trefoil factor 2
|
|
chr15_+_93443559
|
0.066
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chrX_-_13835012
|
0.066
|
|
GPM6B
|
glycoprotein M6B
|
|
chr12_+_28410132
|
0.066
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr10_+_17270257
|
0.065
|
NM_003380
|
VIM
|
vimentin
|
|
chr9_+_130026755
|
0.063
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr22_+_23487512
|
0.062
|
NM_004914
|
RAB36
|
RAB36, member RAS oncogene family
|
|
chr8_+_9953061
|
0.062
|
NM_001135671
NM_001199729
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr21_-_36231794
|
0.062
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr12_-_56756526
|
0.062
|
NM_001638
|
APOF
|
apolipoprotein F
|
|
chr10_+_91152321
|
0.061
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr10_+_123748688
|
0.061
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chrX_+_36065052
|
0.061
|
NM_173695
|
CXorf59
|
chromosome X open reading frame 59
|
|
chr2_+_207406815
|
0.059
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr3_+_136649316
|
0.058
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr9_+_75766794
|
0.058
|
|
ANXA1
|
annexin A1
|
|
chr6_-_26108320
|
0.058
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr18_+_66465316
|
0.057
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr1_-_53608120
|
0.057
|
NM_006671
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7
|
|
chr9_+_12774988
|
0.056
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr1_-_93342706
|
0.056
|
NM_001252271
|
FAM69A
|
family with sequence similarity 69, member A
|
|
chr5_+_140247767
|
0.056
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr3_-_195310985
|
0.056
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr4_+_88754120
|
0.056
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr7_+_102553343
|
0.056
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr11_-_31014232
|
0.055
|
NM_020869
|
DCDC5
|
doublecortin domain containing 5
|
|
chr5_+_96212167
|
0.054
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr4_+_41614911
|
0.054
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_-_64029847
|
0.054
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr11_+_22689653
|
0.053
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr11_+_59856136
|
0.053
|
NM_000139
|
MS4A2
|
membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide)
|
|
chr1_-_150738255
|
0.052
|
|
CTSS
|
cathepsin S
|
|
chr12_+_58003962
|
0.052
|
NM_001111270
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr18_+_28956739
|
0.052
|
NM_001134453
NM_177986
|
DSG4
|
desmoglein 4
|
|
chr14_-_57960432
|
0.052
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr14_+_39583426
|
0.052
|
NM_001009182
NM_001009183
NM_003616
|
GEMIN2
|
gem (nuclear organelle) associated protein 2
|
|
chr16_+_2809933
|
0.052
|
|
|
|
|
chr3_+_194207685
|
0.051
|
|
FLJ34208
|
uncharacterized LOC401106
|
|
chr22_+_44395090
|
0.051
|
NM_001003828
|
PARVB
|
parvin, beta
|
|
chr21_-_36252872
|
0.051
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_171217632
|
0.051
|
NM_002021
|
FMO1
|
flavin containing monooxygenase 1
|
|
chr4_+_74606222
|
0.051
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr7_+_18548899
|
0.050
|
NM_001204147
NM_001204148
|
HDAC9
|
histone deacetylase 9
|
|
chr2_-_134326030
|
0.050
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr3_+_149192367
|
0.050
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr5_-_95158365
|
0.050
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr8_+_98703361
|
0.049
|
|
MTDH
|
metadherin
|
|
chr6_+_139349818
|
0.049
|
NM_021243
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr5_+_36608430
|
0.049
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_-_155774769
|
0.049
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chr1_-_150738232
|
0.049
|
|
CTSS
|
cathepsin S
|
|
chr17_-_39093671
|
0.049
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chrX_+_41548225
|
0.048
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr13_-_34250899
|
0.048
|
NM_001243476
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr8_-_110661007
|
0.048
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr6_-_32908583
|
0.048
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr4_-_23891608
|
0.047
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr3_-_157213138
|
0.047
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr7_+_28338939
|
0.047
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_+_155538096
|
0.046
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr16_-_71610948
|
0.046
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr4_+_71587674
|
0.045
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr1_-_94147201
|
0.045
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr7_+_129015483
|
0.045
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr5_+_140792507
|
0.045
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr10_+_17270469
|
0.045
|
|
VIM
|
vimentin
|
|
chr6_-_47253962
|
0.044
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr13_+_50760015
|
0.044
|
|
|
|
|
chr21_-_43346780
|
0.044
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr5_+_36606667
|
0.043
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr10_-_52008312
|
0.043
|
NM_001143974
NM_019893
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2
|
|
chr10_-_29923900
|
0.043
|
NM_021738
|
SVIL
|
supervillin
|
|
chr2_+_132480063
|
0.042
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr8_-_122653493
|
0.042
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr11_-_124973204
|
0.042
|
NM_001080546
|
TMEM218
|
transmembrane protein 218
|
|
chr6_+_49518112
|
0.042
|
NM_001145652
|
C6orf141
|
chromosome 6 open reading frame 141
|
|
chr2_-_158300517
|
0.042
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chrX_+_100354797
|
0.042
|
NM_006733
|
CENPI
|
centromere protein I
|
|
chrX_+_36254050
|
0.041
|
NM_001098843
|
CXorf30
|
chromosome X open reading frame 30
|
|
chr7_+_107110501
|
0.041
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chrX_-_21776158
|
0.040
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr6_+_25754926
|
0.040
|
NM_005495
|
SLC17A4
|
solute carrier family 17 (sodium phosphate), member 4
|
|
chr8_-_29207795
|
0.040
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr5_+_166711842
|
0.040
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chrX_-_13835313
|
0.039
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_44457424
|
0.039
|
|
CCDC24
|
coiled-coil domain containing 24
|
|
chr9_-_215892
|
0.038
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr6_+_12717036
|
0.038
|
NM_001242648
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr21_-_35899046
|
0.037
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr4_+_169552752
|
0.037
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr1_+_177140062
|
0.037
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr8_+_92114846
|
0.036
|
NM_001129890
|
LRRC69
|
leucine rich repeat containing 69
|
|
chr5_+_150399980
|
0.036
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr3_-_15563160
|
0.036
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr3_-_141747434
|
0.036
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr8_-_102803196
|
0.035
|
|
NCALD
|
neurocalcin delta
|
|
chr8_-_124749573
|
0.035
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr4_+_113218498
|
0.035
|
NM_001102406
NM_025144
|
ALPK1
|
alpha-kinase 1
|
|
chr1_+_244214576
|
0.034
|
|
ZNF238
|
zinc finger protein 238
|
|
chr10_+_35426709
|
0.034
|
NM_183011
NM_183012
|
CREM
|
cAMP responsive element modulator
|
|
chr8_-_62602285
|
0.034
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr7_+_112120907
|
0.034
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr1_-_35450928
|
0.034
|
NM_001195156
|
ZMYM6NB
|
ZMYM6 neighbor
|
|
chr12_+_18414473
|
0.033
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr9_-_112191105
|
0.033
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr17_+_28268622
|
0.033
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr12_+_75784849
|
0.033
|
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr12_-_16757944
|
0.033
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr9_+_130860816
|
0.032
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr19_+_7504573
|
0.032
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr12_+_75784878
|
0.032
|
NM_152436
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chrX_+_100224696
|
0.032
|
NM_001012990
NM_001162490
NM_001162491
|
ARL13A
|
ADP-ribosylation factor-like 13A
|
|
chr3_-_55514933
|
0.031
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr9_-_75567915
|
0.031
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr4_-_10041833
|
0.031
|
NM_001001290
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9
|
|
chr2_+_68961912
|
0.031
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr22_-_26961333
|
0.031
|
NM_001008566
|
TPST2
|
tyrosylprotein sulfotransferase 2
|
|
chr2_+_103378489
|
0.031
|
NM_144632
|
TMEM182
|
transmembrane protein 182
|
|
chr9_+_130860769
|
0.031
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr1_+_207262583
|
0.030
|
NM_000716
NM_001017364
NM_001017367
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr12_+_12223872
|
0.030
|
NM_138723
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator)
|