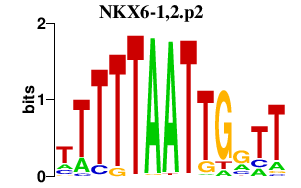
|
chr13_+_27844463
|
0.659
|
NM_206827
|
RASL11A
|
RAS-like, family 11, member A
|
|
chr8_-_82607206
|
0.569
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr6_-_26043884
|
0.491
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr1_-_54483802
|
0.465
|
NM_001010978
|
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1
|
|
chr1_-_150738292
|
0.397
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr11_-_71524890
|
0.397
|
|
LOC285407
|
asparagine-linked glycosylation 1 homolog pseudogene
|
|
chr13_+_27844936
|
0.360
|
|
RASL11A
|
RAS-like, family 11, member A
|
|
chr12_+_32654976
|
0.355
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr13_+_24144402
|
0.339
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr1_+_182419255
|
0.335
|
NM_001137669
|
RGSL1
|
regulator of G-protein signaling like 1
|
|
chr5_+_38445577
|
0.335
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr1_-_221915426
|
0.323
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_-_150738232
|
0.298
|
|
CTSS
|
cathepsin S
|
|
chr5_+_31193852
|
0.297
|
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr1_-_221915393
|
0.295
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr4_+_151503076
|
0.288
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr1_+_186798031
|
0.276
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr7_+_94297448
|
0.275
|
|
PEG10
|
paternally expressed 10
|
|
chr6_+_132859426
|
0.273
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr1_+_168545710
|
0.271
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr1_-_150738255
|
0.254
|
|
CTSS
|
cathepsin S
|
|
chr5_+_31193761
|
0.241
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr12_-_10573041
|
0.239
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr1_+_170115187
|
0.239
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr16_+_28565225
|
0.232
|
NM_138414
|
CCDC101
|
coiled-coil domain containing 101
|
|
chr3_-_138048727
|
0.230
|
NM_178130
|
NME9
|
NME gene family member 9
|
|
chr1_+_157963062
|
0.226
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr7_-_102283237
|
0.218
|
NM_001114403
|
UPK3BL
|
uroplakin 3B-like
|
|
chr21_-_31874407
|
0.215
|
NM_181611
|
KRTAP19-5
|
keratin associated protein 19-5
|
|
chr4_+_74347461
|
0.214
|
NM_001133
|
AFM
|
afamin
|
|
chr2_+_210444364
|
0.210
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr3_-_57234279
|
0.208
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr1_-_221915458
|
0.208
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr6_+_26199712
|
0.205
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr13_+_100229365
|
0.203
|
|
|
|
|
chr21_-_36421195
|
0.200
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_-_70725792
|
0.198
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr21_-_31852631
|
0.194
|
NM_181607
|
KRTAP19-1
|
keratin associated protein 19-1
|
|
chr5_+_149997205
|
0.192
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr5_-_43040147
|
0.192
|
NM_001014279
|
C5orf39
|
chromosome 5 open reading frame 39
|
|
chr2_-_228244005
|
0.183
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr14_-_57960432
|
0.178
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr17_-_67057046
|
0.175
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr2_+_210444154
|
0.169
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr19_-_48759141
|
0.167
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr6_+_53883713
|
0.165
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr4_+_75174189
|
0.165
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr5_+_81601165
|
0.165
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr6_-_146056896
|
0.164
|
NM_001018041
NM_005670
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin)
|
|
chr18_-_51690988
|
0.163
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr18_-_46477080
|
0.161
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr12_+_54410893
|
0.159
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr4_-_170077807
|
0.159
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr20_-_18477811
|
0.158
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr13_-_47470967
|
0.155
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr17_+_68100994
|
0.154
|
NM_170742
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr7_-_92855780
|
0.148
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr21_-_36421578
|
0.148
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_-_135271206
|
0.148
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr11_-_125550674
|
0.148
|
NM_001612
NM_020069
NM_020107
NM_020108
|
ACRV1
|
acrosomal vesicle protein 1
|
|
chr4_+_88896899
|
0.147
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr7_+_28725597
|
0.146
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_-_16760935
|
0.146
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_-_79817058
|
0.144
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chrX_-_33229428
|
0.138
|
NM_004006
|
DMD
|
dystrophin
|
|
chr6_+_5030202
|
0.138
|
|
LOC100129461
|
uncharacterized LOC100129461
|
|
chr8_+_79428335
|
0.135
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr4_+_74606222
|
0.134
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr5_-_135290521
|
0.134
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr11_-_124190092
|
0.132
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr19_-_41812991
|
0.131
|
|
|
|
|
chr1_-_186430216
|
0.129
|
NM_002597
|
PDC
|
phosducin
|
|
chr2_+_211342405
|
0.127
|
NM_001122633
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr4_-_70518964
|
0.126
|
NM_001252274
NM_001252275
NM_006798
|
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus
|
|
chr3_-_151987332
|
0.126
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr1_-_17766142
|
0.125
|
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr1_-_173020011
|
0.124
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr7_+_128312345
|
0.123
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr13_+_108922560
|
0.122
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr11_-_67205151
|
0.121
|
NM_005608
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein
|
|
chr21_-_36421461
|
0.121
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_+_94750041
|
0.120
|
NM_005172
|
ATOH1
|
atonal homolog 1 (Drosophila)
|
|
chr5_+_140566655
|
0.120
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr16_-_3843474
|
0.119
|
|
CREBBP
|
CREB binding protein
|
|
chr6_+_39760138
|
0.119
|
|
|
|
|
chr18_-_24722757
|
0.117
|
NM_001243848
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr1_-_186417855
|
0.114
|
NM_022576
|
PDC
|
phosducin
|
|
chr17_+_68071386
|
0.114
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr7_+_64601439
|
0.114
|
|
INTS4L1
|
integrator complex subunit 4-like 1
|
|
chr14_+_74083547
|
0.113
|
NM_001037162
|
ACOT6
|
acyl-CoA thioesterase 6
|
|
chrX_+_41548225
|
0.112
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr7_-_20256942
|
0.112
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr3_-_71294229
|
0.111
|
NM_001244814
|
FOXP1
|
forkhead box P1
|
|
chr10_-_96829074
|
0.109
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr6_-_133084585
|
0.109
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr2_-_157186681
|
0.109
|
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr1_+_210501595
|
0.107
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr7_-_41742666
|
0.107
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr12_-_109458844
|
0.107
|
NM_018711
|
SVOP
|
SV2 related protein homolog (rat)
|
|
chr1_+_67773046
|
0.107
|
NM_001559
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chr16_-_3627353
|
0.106
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr10_+_72163899
|
0.105
|
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr2_-_74382352
|
0.104
|
|
|
|
|
chr4_+_15704752
|
0.102
|
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr8_+_132991171
|
0.101
|
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr12_+_25205488
|
0.101
|
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr3_+_51575595
|
0.099
|
NM_015106
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae)
|
|
chr1_+_61869699
|
0.099
|
|
NFIA
|
nuclear factor I/A
|
|
chr15_+_34261088
|
0.099
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr16_-_46782104
|
0.098
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr2_-_151344145
|
0.095
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr2_-_175712276
|
0.095
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr11_-_62521533
|
0.095
|
NM_024784
|
ZBTB3
|
zinc finger and BTB domain containing 3
|
|
chr1_-_17766241
|
0.095
|
NM_018715
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr6_-_138820580
|
0.094
|
|
|
|
|
chr21_-_36421630
|
0.094
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr19_+_9296269
|
0.094
|
NM_175883
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2
|
|
chr11_+_12308446
|
0.093
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr12_-_53558276
|
0.093
|
NM_001244706
|
CSAD
|
cysteine sulfinic acid decarboxylase
|
|
chr4_+_155484131
|
0.092
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr16_+_69680959
|
0.091
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr14_-_21567069
|
0.091
|
NM_001101672
|
ZNF219
|
zinc finger protein 219
|
|
chr2_+_29038862
|
0.089
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr12_-_46357949
|
0.088
|
|
|
|
|
chr6_-_138893666
|
0.087
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr17_-_7307413
|
0.087
|
NM_152766
|
C17orf61-PLSCR3
C17orf61
|
C17orf61-PLSCR3 readthrough
chromosome 17 open reading frame 61
|
|
chr19_-_3029611
|
0.086
|
NM_001144762
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr10_-_94257470
|
0.086
|
NM_001165946
|
IDE
|
insulin-degrading enzyme
|
|
chr13_-_44203608
|
0.086
|
NM_001127615
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr3_-_119582422
|
0.086
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr1_-_6420719
|
0.085
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr6_-_138820578
|
0.084
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr6_-_49712055
|
0.084
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr21_-_36421586
|
0.083
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr7_+_137761177
|
0.083
|
NM_001190906
NM_001190907
NM_005989
|
AKR1D1
|
aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase)
|
|
chr2_+_143635225
|
0.083
|
|
KYNU
|
kynureninase
|
|
chr4_-_17806820
|
0.082
|
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr14_-_24657986
|
0.082
|
|
IPO4
|
importin 4
|
|
chr2_-_202563353
|
0.082
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr3_+_156394452
|
0.081
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr19_+_24269877
|
0.080
|
NM_203282
|
ZNF254
|
zinc finger protein 254
|
|
chr8_+_9953061
|
0.080
|
NM_001135671
NM_001199729
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr19_+_1270926
|
0.079
|
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr17_+_28268622
|
0.078
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr14_-_24658010
|
0.077
|
NM_024658
|
IPO4
|
importin 4
|
|
chr9_-_73483933
|
0.077
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr13_+_99058147
|
0.077
|
|
|
|
|
chr12_+_56732658
|
0.077
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr4_-_185726865
|
0.077
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr1_+_74701070
|
0.077
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr20_+_23345020
|
0.076
|
NM_022482
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr3_-_47957475
|
0.076
|
|
|
|
|
chr16_-_72128214
|
0.076
|
NM_001142318
|
TXNL4B
|
thioredoxin-like 4B
|
|
chr22_-_19435727
|
0.074
|
NM_001166242
NM_173793
|
C22orf39
|
chromosome 22 open reading frame 39
|
|
chr5_+_145316120
|
0.072
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr10_+_114043413
|
0.072
|
NM_058222
|
TECTB
|
tectorin beta
|
|
chr7_+_143657019
|
0.071
|
NM_012369
|
OR2F1
|
olfactory receptor, family 2, subfamily F, member 1
|
|
chr20_+_19915716
|
0.070
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr17_+_38975343
|
0.070
|
NM_001195387
NM_145274
NM_001195386
|
TMEM99
|
transmembrane protein 99
|
|
chrX_-_24690740
|
0.070
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr20_+_51801821
|
0.070
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr7_-_92848837
|
0.070
|
NM_198151
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr2_+_183608329
|
0.070
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr7_+_55433140
|
0.069
|
NM_018697
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial)
|
|
chr12_-_54653317
|
0.069
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr3_+_49547867
|
0.068
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr6_+_114178516
|
0.068
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr7_+_120965420
|
0.067
|
NM_016087
|
WNT16
|
wingless-type MMTV integration site family, member 16
|
|
chr1_+_174844655
|
0.066
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr7_+_72396799
|
0.065
|
|
POM121
|
POM121 membrane glycoprotein
|
|
chr12_-_54653369
|
0.065
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr7_+_143880547
|
0.064
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr9_-_73736513
|
0.064
|
NM_001007471
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr1_-_149858114
|
0.062
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr22_-_22863504
|
0.062
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr8_-_72268720
|
0.062
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr16_+_67311412
|
0.061
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr6_+_159070994
|
0.060
|
NM_001009991
NM_001242384
NM_001242394
NM_001242395
|
SYTL3
|
synaptotagmin-like 3
|
|
chr2_+_143635194
|
0.060
|
NM_001032998
NM_001199241
NM_003937
|
KYNU
|
kynureninase
|
|
chr6_-_138539626
|
0.060
|
NM_021635
|
PBOV1
|
prostate and breast cancer overexpressed 1
|
|
chr12_-_53994783
|
0.060
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr16_-_54962707
|
0.060
|
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr4_+_88896801
|
0.060
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chrM_+_14601
|
0.059
|
|
|
|
|
chr16_+_70333058
|
0.059
|
NM_001014449
NM_001014451
NM_007242
|
DDX19B
|
DEAD (Asp-Glu-Ala-As) box polypeptide 19B
|
|
chr5_+_140800536
|
0.058
|
NM_018914
NM_032091
NM_032092
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr4_+_169418167
|
0.057
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr10_+_63808968
|
0.057
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr17_+_76000248
|
0.057
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr19_+_45394522
|
0.057
|
|
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr15_+_83477972
|
0.057
|
NM_001080435
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules
|
|
chr8_-_13372183
|
0.057
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr12_-_56679729
|
0.057
|
|
CS
|
citrate synthase
|
|
chr2_+_108994366
|
0.056
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr3_+_49506079
|
0.056
|
NM_001177643
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr3_-_126327397
|
0.056
|
NM_001039783
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor
|
|
chr17_-_56494930
|
0.056
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chrX_+_107288199
|
0.056
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr5_+_140749830
|
0.055
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr13_+_97928414
|
0.055
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|