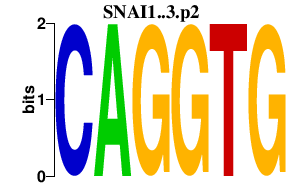
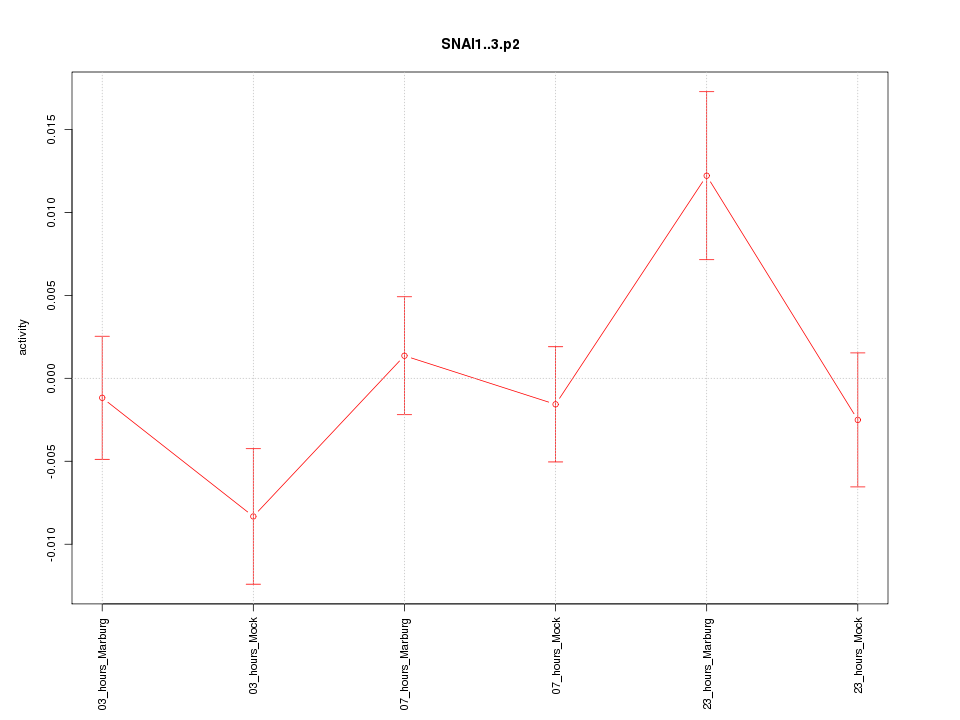
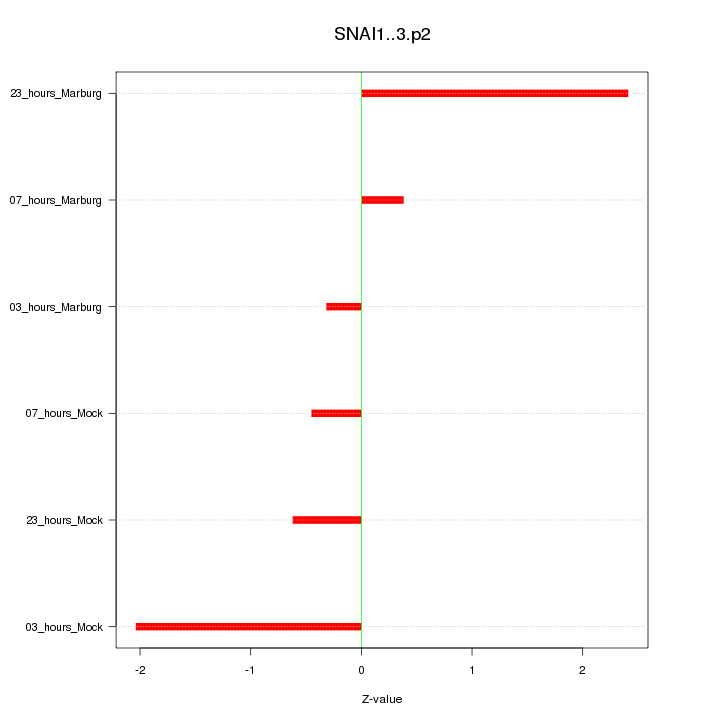
|
chr19_-_55836668
|
1.794
|
NM_001085488
|
TMEM150B
|
transmembrane protein 150B
|
|
chr17_+_43299277
|
1.747
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr19_-_12886263
|
1.742
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr8_+_32405727
|
1.544
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr6_-_136871542
|
1.469
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr6_+_36646489
|
1.261
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr13_-_21295739
|
1.256
|
|
|
|
|
chr11_-_560706
|
1.253
|
NM_173573
|
C11orf35
|
chromosome 11 open reading frame 35
|
|
chr1_-_40367539
|
1.235
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr11_+_111789600
|
1.198
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr7_+_140103842
|
1.185
|
NM_001008749
|
RAB19
|
RAB19, member RAS oncogene family
|
|
chr1_-_85514174
|
1.149
|
NM_001253693
NM_018298
|
MCOLN3
|
mucolipin 3
|
|
chr21_+_37507209
|
1.138
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr1_-_27286875
|
1.115
|
NM_152365
|
C1orf172
|
chromosome 1 open reading frame 172
|
|
chr1_-_55266870
|
1.114
|
NM_001114108
NM_017904
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chr8_-_17658711
|
1.092
|
|
|
|
|
chr5_+_148520985
|
1.077
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr1_-_85462622
|
1.071
|
NM_153259
|
MCOLN2
|
mucolipin 2
|
|
chr6_+_36646455
|
0.992
|
NM_000389
NM_001220778
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr7_+_156803599
|
0.982
|
|
LOC645249
|
uncharacterized LOC645249
|
|
chr2_+_121103670
|
0.964
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr8_+_22436641
|
0.951
|
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr5_-_43412417
|
0.944
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr8_+_99076749
|
0.931
|
NM_001170806
NM_173549
|
C8orf47
|
chromosome 8 open reading frame 47
|
|
chr19_-_12886421
|
0.927
|
NM_001100176
NM_013312
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr16_+_83986826
|
0.925
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr5_+_71403040
|
0.911
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr11_+_111789658
|
0.905
|
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr5_-_138718941
|
0.890
|
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr11_+_77774902
|
0.880
|
NM_003251
|
THRSP
|
thyroid hormone responsive
|
|
chr15_+_41136622
|
0.870
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr6_+_36098261
|
0.866
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr2_-_166650770
|
0.864
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr6_-_132722564
|
0.844
|
NM_015529
|
MOXD1
|
monooxygenase, DBH-like 1
|
|
chr8_+_21882353
|
0.844
|
NM_182795
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr5_+_76114832
|
0.841
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr1_+_177140530
|
0.825
|
NM_021165
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr22_-_24096551
|
0.814
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr4_-_25864430
|
0.809
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr8_+_21881587
|
0.797
|
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr2_+_210288668
|
0.794
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr1_-_111746917
|
0.768
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr9_+_116111811
|
0.766
|
NM_017688
|
BSPRY
|
B-box and SPRY domain containing
|
|
chr15_+_41136178
|
0.762
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr6_+_30130982
|
0.742
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr7_+_12610202
|
0.739
|
NM_001112706
|
SCIN
|
scinderin
|
|
chr6_+_22569656
|
0.736
|
NM_138574
|
HDGFL1
|
hepatoma derived growth factor-like 1
|
|
chr6_+_33589045
|
0.736
|
NM_002224
|
ITPR3
|
inositol 1,4,5-trisphosphate receptor, type 3
|
|
chr8_-_119634116
|
0.735
|
|
SAMD12
|
sterile alpha motif domain containing 12
|
|
chr16_+_3062456
|
0.732
|
NM_020982
|
CLDN9
|
claudin 9
|
|
chr7_+_130020289
|
0.717
|
NM_001868
|
CPA1
|
carboxypeptidase A1 (pancreatic)
|
|
chr11_+_71259465
|
0.713
|
NM_005553
|
KRTAP5-9
|
keratin associated protein 5-9
|
|
chr1_-_183559463
|
0.688
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr6_+_54711568
|
0.688
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr3_+_119421865
|
0.687
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr1_-_85514128
|
0.686
|
|
MCOLN3
|
mucolipin 3
|
|
chr1_+_46646199
|
0.679
|
|
TSPAN1
|
tetraspanin 1
|
|
chr5_+_76114881
|
0.676
|
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr19_-_19738973
|
0.673
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr17_-_7165788
|
0.671
|
|
CLDN7
|
claudin 7
|
|
chr8_+_22436253
|
0.667
|
NM_021630
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr14_+_75745480
|
0.653
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr17_-_40575117
|
0.650
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr16_+_71660063
|
0.639
|
NM_001017967
NM_052858
|
MARVELD3
|
MARVEL domain containing 3
|
|
chr16_+_68771214
|
0.639
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr1_+_183155393
|
0.634
|
|
LAMC2
|
laminin, gamma 2
|
|
chr1_+_239549834
|
0.624
|
|
CHRM3
|
cholinergic receptor, muscarinic 3
|
|
chr4_+_85504056
|
0.624
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chrX_-_21676416
|
0.622
|
NM_153270
|
KLHL34
|
kelch-like 34 (Drosophila)
|
|
chr12_-_26277807
|
0.620
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr3_-_8693723
|
0.618
|
NM_015931
|
C3orf32
|
chromosome 3 open reading frame 32
|
|
chr19_-_36499583
|
0.615
|
NM_001039876
|
C19orf46
|
chromosome 19 open reading frame 46
|
|
chr4_-_74486100
|
0.613
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr5_-_138719018
|
0.607
|
NM_005847
NM_152685
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr17_+_27920479
|
0.602
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr4_-_139163222
|
0.602
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr5_+_59784004
|
0.601
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr3_+_67705120
|
0.597
|
|
|
|
|
chr3_-_9920471
|
0.592
|
NM_001199623
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr7_+_134212343
|
0.589
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr2_-_166650705
|
0.585
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr2_+_112813079
|
0.585
|
|
TMEM87B
|
transmembrane protein 87B
|
|
chr16_+_330605
|
0.585
|
NM_001176
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma
|
|
chr19_+_38794800
|
0.581
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr1_+_19934573
|
0.575
|
|
|
|
|
chr16_+_2039945
|
0.575
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr8_+_87111132
|
0.572
|
NM_152565
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2
|
|
chr16_-_75529278
|
0.566
|
|
|
|
|
chr6_+_37137882
|
0.563
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr14_-_65409445
|
0.563
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr5_+_87564840
|
0.562
|
|
LOC100505894
|
uncharacterized LOC100505894
|
|
chr10_-_62493077
|
0.562
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr11_+_65554566
|
0.562
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr6_+_31783314
|
0.561
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr2_+_112812900
|
0.560
|
|
TMEM87B
|
transmembrane protein 87B
|
|
chr2_+_27346656
|
0.559
|
NM_032604
|
ABHD1
|
abhydrolase domain containing 1
|
|
chr1_+_177140062
|
0.558
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr16_-_30107492
|
0.557
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr17_-_43319100
|
0.557
|
|
|
|
|
chr1_-_51810693
|
0.556
|
NM_001144832
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr6_+_17282931
|
0.554
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr16_+_68771192
|
0.552
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr8_+_22437983
|
0.549
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr6_+_31783530
|
0.549
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr4_-_2420252
|
0.547
|
NM_001172656
NM_001172657
NM_001172658
NM_020972
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr13_+_44947977
|
0.544
|
NM_001010897
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2
|
|
chr10_-_28591844
|
0.543
|
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7)
|
|
chr2_+_112812799
|
0.540
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr7_-_107880492
|
0.539
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr1_-_201368617
|
0.539
|
NM_005558
|
LAD1
|
ladinin 1
|
|
chr5_-_172756505
|
0.537
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr16_-_4987056
|
0.537
|
NM_002705
|
PPL
|
periplakin
|
|
chr12_-_51785192
|
0.537
|
NM_007210
|
GALNT6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6)
|
|
chr9_+_140119643
|
0.536
|
NM_199001
|
C9orf169
|
chromosome 9 open reading frame 169
|
|
chr22_-_38506532
|
0.536
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr7_-_87936202
|
0.536
|
NM_001205315
NM_001205316
NM_024636
|
STEAP4
|
STEAP family member 4
|
|
chr1_-_1590403
|
0.534
|
|
CDK11A
CDK11B
|
cyclin-dependent kinase 11A
cyclin-dependent kinase 11B
|
|
chr1_+_1981902
|
0.533
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr7_-_139168408
|
0.532
|
NM_198508
|
KLRG2
|
killer cell lectin-like receptor subfamily G, member 2
|
|
chr7_+_44078677
|
0.530
|
|
FLJ35390
|
uncharacterized LOC255031
|
|
chr11_+_59522801
|
0.528
|
|
STX3
|
syntaxin 3
|
|
chr14_-_105647520
|
0.528
|
NM_177533
|
NUDT14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14
|
|
chr12_+_109551041
|
0.527
|
|
|
|
|
chr6_-_46293215
|
0.525
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr1_+_183155361
|
0.525
|
|
LAMC2
|
laminin, gamma 2
|
|
chr11_+_32851469
|
0.522
|
NM_024081
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr15_+_50474357
|
0.521
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr7_-_143892649
|
0.518
|
NM_001003702
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chr1_+_145524989
|
0.516
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr9_+_92219926
|
0.515
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr3_-_57199402
|
0.511
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr3_+_32023265
|
0.510
|
NM_001137674
|
ZNF860
|
zinc finger protein 860
|
|
chr15_-_60884615
|
0.510
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr7_+_37960162
|
0.509
|
NM_001242946
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr11_+_130029773
|
0.509
|
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma)
|
|
chr16_+_24857551
|
0.508
|
NM_052944
|
SLC5A11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11
|
|
chr14_+_24600674
|
0.507
|
NM_203402
|
FITM1
|
fat storage-inducing transmembrane protein 1
|
|
chr1_+_32827797
|
0.506
|
NM_052841
|
TSSK3
|
testis-specific serine kinase 3
|
|
chr16_+_68771236
|
0.505
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr16_+_28303622
|
0.503
|
NM_001024401
|
SBK1
|
SH3-binding domain kinase 1
|
|
chr16_+_68771304
|
0.501
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr3_-_133614296
|
0.500
|
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr19_+_14017038
|
0.495
|
|
CC2D1A
|
coiled-coil and C2 domain containing 1A
|
|
chr6_-_46459010
|
0.494
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chr1_-_147931877
|
0.492
|
|
FLJ39739
|
uncharacterized FLJ39739
|
|
chr12_-_53600999
|
0.491
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr11_+_10476665
|
0.491
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr1_-_160990971
|
0.489
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr20_+_48599511
|
0.486
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr2_-_238499713
|
0.486
|
NM_022449
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr2_-_28113211
|
0.484
|
NM_022128
|
RBKS
|
ribokinase
|
|
chr8_-_6420454
|
0.483
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr17_+_42836546
|
0.483
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr15_+_45927217
|
0.483
|
NM_021199
|
SQRDL
|
sulfide quinone reductase-like (yeast)
|
|
chr19_+_6531009
|
0.481
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr17_-_72968790
|
0.479
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr19_-_49622278
|
0.476
|
NM_018111
|
C19orf73
|
chromosome 19 open reading frame 73
|
|
chr7_-_22396511
|
0.474
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr17_-_73874056
|
0.473
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr4_-_101111606
|
0.473
|
NM_145244
|
DDIT4L
|
DNA-damage-inducible transcript 4-like
|
|
chr17_-_31204071
|
0.472
|
|
MYO1D
|
myosin ID
|
|
chr4_+_152330326
|
0.470
|
NM_001109977
|
FAM160A1
|
family with sequence similarity 160, member A1
|
|
chr18_-_47721165
|
0.470
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr9_+_114287438
|
0.470
|
NM_001007169
NM_133464
|
ZNF483
|
zinc finger protein 483
|
|
chr1_-_201368434
|
0.469
|
|
LAD1
|
ladinin 1
|
|
chr8_-_144815888
|
0.468
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr16_+_81812899
|
0.468
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr5_-_75919051
|
0.468
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr1_-_24741586
|
0.466
|
NM_001199012
|
C1orf201
|
chromosome 1 open reading frame 201
|
|
chr2_-_238499317
|
0.460
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr1_-_40782826
|
0.457
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr12_-_105478218
|
0.455
|
NM_001034173
|
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2
|
|
chr17_-_43502992
|
0.455
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr8_+_143808593
|
0.453
|
NM_016647
|
C8orf55
|
chromosome 8 open reading frame 55
|
|
chr15_-_41166336
|
0.453
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr16_-_850698
|
0.452
|
NM_016541
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13
|
|
chr7_+_29603499
|
0.451
|
|
PRR15
|
proline rich 15
|
|
chr7_-_124405029
|
0.447
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr2_-_122042767
|
0.445
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr3_-_32023237
|
0.445
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr1_-_183559669
|
0.444
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr6_+_31795395
|
0.442
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr15_-_42186274
|
0.441
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr17_+_76126835
|
0.441
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr14_-_51561657
|
0.441
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr2_+_219745243
|
0.439
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr17_-_31203889
|
0.437
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr11_+_65554504
|
0.435
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr12_-_39299335
|
0.435
|
NM_153634
|
CPNE8
|
copine VIII
|
|
chr12_+_6309475
|
0.435
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr17_-_39684499
|
0.434
|
|
KRT19
|
keratin 19
|
|
chr7_-_97601654
|
0.434
|
|
|
|
|
chr9_+_2621740
|
0.434
|
NM_001018056
NM_003383
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr16_-_3068161
|
0.433
|
NM_021195
|
CLDN6
|
claudin 6
|
|
chr1_-_144520918
|
0.433
|
|
LOC728855
|
uncharacterized LOC728855
|
|
chr18_-_74844710
|
0.432
|
|
MBP
|
myelin basic protein
|
|
chr1_-_168513201
|
0.432
|
NM_003175
|
XCL1
XCL2
|
chemokine (C motif) ligand 1
chemokine (C motif) ligand 2
|
|
chr19_+_14017044
|
0.431
|
|
CC2D1A
|
coiled-coil and C2 domain containing 1A
|
|
chr10_-_62493282
|
0.430
|
NM_001204403
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr2_-_26205327
|
0.429
|
|
KIF3C
|
kinesin family member 3C
|