|
chr9_+_92219926
|
1.961
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr2_-_161056762
|
1.127
|
|
ITGB6
|
integrin, beta 6
|
|
chr1_-_89530890
|
0.990
|
NM_002053
|
GBP1
|
guanylate binding protein 1, interferon-inducible
|
|
chr17_+_57915192
|
0.942
|
|
|
|
|
chr17_+_3379295
|
0.789
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr20_+_35169876
|
0.784
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr10_-_29923900
|
0.781
|
NM_021738
|
SVIL
|
supervillin
|
|
chr20_+_35169897
|
0.743
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr1_-_11907741
|
0.728
|
NM_006172
|
NPPA
|
natriuretic peptide A
|
|
chr12_-_50616425
|
0.682
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr20_+_2276612
|
0.670
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr2_-_128432642
|
0.666
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr2_-_96810775
|
0.662
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr6_-_29527701
|
0.607
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr2_-_96811138
|
0.579
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr5_+_137801178
|
0.546
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr2_-_106015423
|
0.536
|
NM_001039492
NM_001450
NM_201555
|
FHL2
|
four and a half LIM domains 2
|
|
chr2_-_161056573
|
0.531
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr20_-_656443
|
0.523
|
|
|
|
|
chr12_+_75874512
|
0.472
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr6_+_151561094
|
0.470
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr12_+_26348488
|
0.469
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr19_+_16178170
|
0.433
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr13_-_46742678
|
0.424
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr11_-_65667809
|
0.416
|
|
FOSL1
|
FOS-like antigen 1
|
|
chr1_-_168106810
|
0.414
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr10_-_64576074
|
0.401
|
|
EGR2
|
early growth response 2
|
|
chr3_-_123339423
|
0.390
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr9_+_116356199
|
0.387
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr5_-_33891967
|
0.381
|
NM_030955
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr4_+_6695565
|
0.368
|
NM_005980
|
S100P
|
S100 calcium binding protein P
|
|
chr10_+_88428263
|
0.363
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr17_+_16318848
|
0.354
|
NM_016113
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr3_-_123339178
|
0.352
|
|
MYLK
|
myosin light chain kinase
|
|
chr22_+_31480981
|
0.350
|
NM_001207018
|
SMTN
|
smoothelin
|
|
chr17_-_79479831
|
0.348
|
NM_001199954
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chrX_-_153602928
|
0.346
|
|
FLNA
|
filamin A, alpha
|
|
chr6_-_29527477
|
0.335
|
|
UBD
|
ubiquitin D
|
|
chr5_-_33892292
|
0.334
|
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr1_-_168106535
|
0.333
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr17_-_4890920
|
0.326
|
NM_001171166
NM_015099
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr1_+_167599503
|
0.325
|
|
RCSD1
|
RCSD domain containing 1
|
|
chr11_-_65667860
|
0.317
|
NM_005438
|
FOSL1
|
FOS-like antigen 1
|
|
chr1_-_168106727
|
0.311
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr19_+_1026608
|
0.300
|
|
CNN2
|
calponin 2
|
|
chr7_-_27183225
|
0.299
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr1_-_935469
|
0.287
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr6_+_151561508
|
0.286
|
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr6_-_109775423
|
0.283
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr7_+_143078358
|
0.283
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr9_+_139249251
|
0.278
|
NM_001200003
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr6_-_105584982
|
0.278
|
NM_001199563
|
BVES
|
blood vessel epicardial substance
|
|
chr19_+_45971249
|
0.277
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr7_-_559479
|
0.275
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr19_+_1026624
|
0.268
|
|
CNN2
|
calponin 2
|
|
chr17_-_79479780
|
0.265
|
|
ACTG1
|
actin, gamma 1
|
|
chr7_+_143078395
|
0.263
|
|
ZYX
|
zyxin
|
|
chr17_+_16318911
|
0.263
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr17_-_39684585
|
0.263
|
NM_002276
|
KRT19
|
keratin 19
|
|
chr14_+_75745480
|
0.256
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr7_+_143078444
|
0.251
|
|
ZYX
|
zyxin
|
|
chr4_+_89299874
|
0.242
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr5_-_53606387
|
0.241
|
NM_019087
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr1_+_16085245
|
0.241
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr4_-_10118460
|
0.239
|
|
WDR1
|
WD repeat domain 1
|
|
chr9_+_36136710
|
0.238
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr7_+_143078399
|
0.234
|
|
ZYX
|
zyxin
|
|
chr10_-_16859336
|
0.223
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr4_+_108814419
|
0.221
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr11_+_66188474
|
0.220
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chr4_+_24797084
|
0.219
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr10_+_112257624
|
0.216
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr10_-_16859408
|
0.215
|
NM_012425
NM_152724
|
RSU1
|
Ras suppressor protein 1
|
|
chrX_-_153602998
|
0.211
|
NM_001110556
NM_001456
|
FLNA
|
filamin A, alpha
|
|
chr1_+_223889253
|
0.208
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr3_-_99594915
|
0.208
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr10_-_16859389
|
0.207
|
|
RSU1
|
Ras suppressor protein 1
|
|
chr10_-_64576123
|
0.204
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr19_+_1026627
|
0.202
|
|
CNN2
|
calponin 2
|
|
chr7_+_143078447
|
0.201
|
|
ZYX
|
zyxin
|
|
chr7_+_143078457
|
0.199
|
|
ZYX
|
zyxin
|
|
chr12_-_57443858
|
0.195
|
NM_005379
|
MYO1A
|
myosin IA
|
|
chr4_-_10118444
|
0.193
|
|
WDR1
|
WD repeat domain 1
|
|
chr12_+_52445185
|
0.193
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr1_-_89591685
|
0.193
|
NM_004120
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chr19_+_1026582
|
0.189
|
|
CNN2
|
calponin 2
|
|
chr19_-_45826131
|
0.181
|
|
CKM
|
creatine kinase, muscle
|
|
chr2_+_219283809
|
0.178
|
NM_007127
|
VIL1
|
villin 1
|
|
chr9_+_116356407
|
0.178
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr5_-_53606303
|
0.172
|
|
ARL15
|
ADP-ribosylation factor-like 15
|
|
chr10_-_131762374
|
0.168
|
|
EBF3
|
early B-cell factor 3
|
|
chr12_+_50497790
|
0.165
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr7_+_143078608
|
0.165
|
|
ZYX
|
zyxin
|
|
chr2_+_74120132
|
0.161
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr6_-_16761600
|
0.160
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr19_+_54372659
|
0.154
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr17_-_4890664
|
0.149
|
NM_001171167
NM_001171168
|
CAMTA2
|
calmodulin binding transcription activator 2
|
|
chr6_-_76203256
|
0.147
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr19_+_16187306
|
0.143
|
|
TPM4
|
tropomyosin 4
|
|
chr19_-_36247917
|
0.140
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr19_+_56153417
|
0.139
|
NM_016202
NM_001163423
|
ZNF580
|
zinc finger protein 580
|
|
chr8_-_22550708
|
0.137
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr1_+_50574589
|
0.137
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr15_-_55581969
|
0.134
|
NM_183235
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr19_+_49376488
|
0.133
|
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr17_-_3867585
|
0.130
|
NM_005173
NM_174953
NM_174954
NM_174955
NM_174956
NM_174957
NM_174958
|
ATP2A3
|
ATPase, Ca++ transporting, ubiquitous
|
|
chr7_+_115863004
|
0.122
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr13_+_27825689
|
0.122
|
NM_000982
|
RPL21
|
ribosomal protein L21
|
|
chrX_-_153599719
|
0.118
|
|
FLNA
|
filamin A, alpha
|
|
chr19_+_48958765
|
0.117
|
NM_013348
|
KCNJ14
|
potassium inwardly-rectifying channel, subfamily J, member 14
|
|
chr7_-_5570188
|
0.115
|
NM_001101
|
ACTB
|
actin, beta
|
|
chr10_+_75757847
|
0.114
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chr10_-_82049178
|
0.114
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr6_+_43139201
|
0.113
|
|
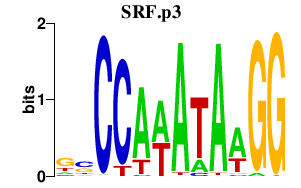
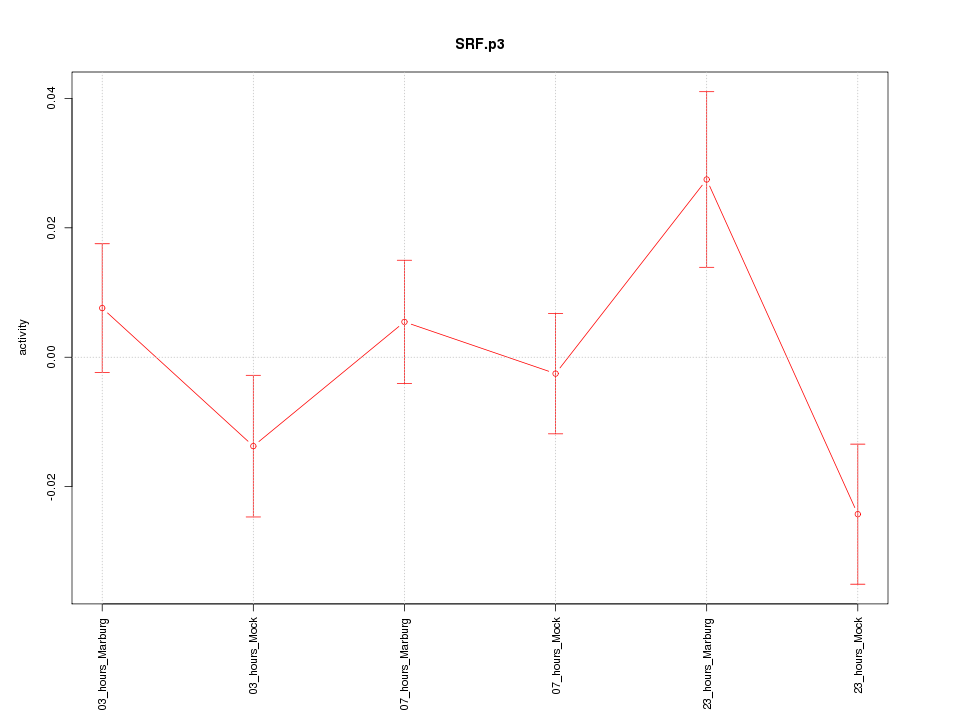
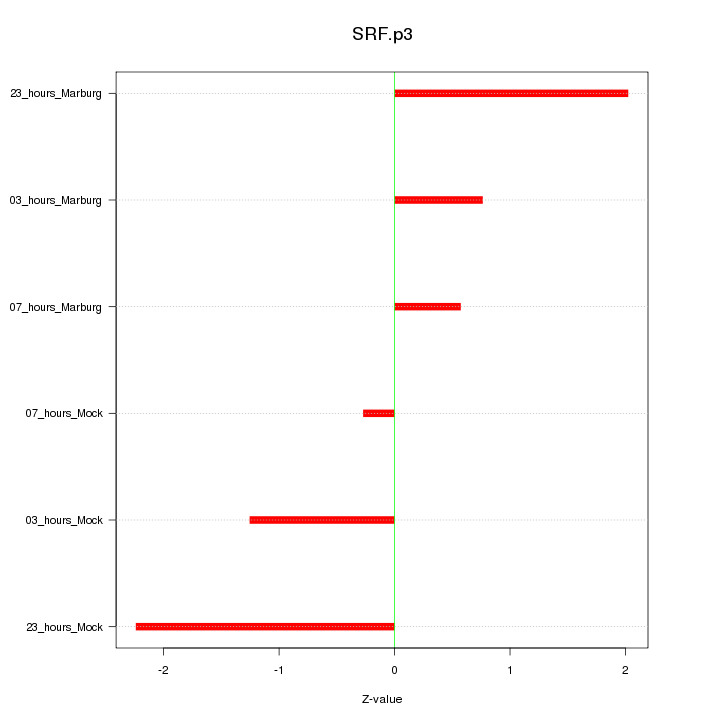
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr19_+_57874878
|
0.113
|
NM_173631
|
TRAPPC2P1
ZNF547
|
trafficking protein particle complex 2 pseudogene 1
zinc finger protein 547
|
|
chr19_+_47813074
|
0.112
|
NM_001736
|
C5AR1
|
complement component 5a receptor 1
|
|
chr4_-_10118500
|
0.108
|
NM_005112
NM_017491
|
WDR1
|
WD repeat domain 1
|
|
chrX_-_10851808
|
0.099
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr9_-_98269686
|
0.098
|
NM_001083606
|
PTCH1
|
patched 1
|
|
chr1_-_229569839
|
0.094
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr11_-_30605748
|
0.093
|
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr19_+_16187047
|
0.092
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr3_-_9834357
|
0.090
|
NM_006354
NM_133480
|
TADA3
|
transcriptional adaptor 3
|
|
chr1_-_935352
|
0.089
|
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr1_+_181057835
|
0.088
|
|
IER5
|
immediate early response 5
|
|
chr5_+_176875325
|
0.086
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr14_+_105267378
|
0.085
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr17_+_41003184
|
0.084
|
NM_003734
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr1_-_150552077
|
0.084
|
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr15_-_59665069
|
0.082
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr17_-_1389323
|
0.082
|
|
MYO1C
|
myosin IC
|
|
chr15_+_71433787
|
0.081
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr6_+_43139190
|
0.078
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr19_-_15343767
|
0.073
|
NM_001142886
|
EPHX3
|
epoxide hydrolase 3
|
|
chr6_+_126240369
|
0.073
|
NM_001199622
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr17_-_10372875
|
0.070
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr17_-_41132019
|
0.069
|
NM_025267
|
AARSD1
|
alanyl-tRNA synthetase domain containing 1
|
|
chr10_+_123922940
|
0.068
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr12_+_8276227
|
0.064
|
NM_016184
NM_194447
NM_194448
NM_194450
|
CLEC4A
|
C-type lectin domain family 4, member A
|
|
chr3_+_143690639
|
0.063
|
NM_173552
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr14_+_78227172
|
0.062
|
NM_001173978
NM_174943
|
C14orf178
|
chromosome 14 open reading frame 178
|
|
chr2_+_87754991
|
0.061
|
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr2_+_87755059
|
0.061
|
|
LINC00152
|
long intergenic non-protein coding RNA 152
|
|
chr15_-_59665065
|
0.061
|
|
MYO1E
|
myosin IE
|
|
chr17_+_33474835
|
0.060
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr3_+_25470067
|
0.060
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr3_+_49027281
|
0.060
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr19_-_12792676
|
0.058
|
|
DHPS
|
deoxyhypusine synthase
|
|
chr3_+_187930720
|
0.057
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr18_-_3219846
|
0.057
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr6_+_158402887
|
0.057
|
NM_003898
|
SYNJ2
|
synaptojanin 2
|
|
chr5_-_58295758
|
0.057
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_140749830
|
0.056
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr7_-_107643330
|
0.056
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr7_+_93551015
|
0.056
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr19_+_1026271
|
0.054
|
NM_004368
NM_201277
|
CNN2
|
calponin 2
|
|
chr22_-_36784055
|
0.053
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr21_-_28217272
|
0.053
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr9_-_98269480
|
0.051
|
NM_001083607
|
PTCH1
|
patched 1
|
|
chr3_-_151987332
|
0.051
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr18_+_29598444
|
0.051
|
NM_017831
|
RNF125
|
ring finger protein 125
|
|
chr20_-_42815732
|
0.050
|
|
JPH2
|
junctophilin 2
|
|
chr15_+_45406518
|
0.048
|
NM_207581
|
DUOXA2
|
dual oxidase maturation factor 2
|
|
chr4_-_40632639
|
0.048
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr7_+_134464161
|
0.048
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr5_-_39425297
|
0.048
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr1_-_154164551
|
0.046
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr1_-_217113007
|
0.046
|
NM_001243512
NM_001243513
NM_001243510
NM_001243511
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr4_+_77227882
|
0.045
|
|
STBD1
|
starch binding domain 1
|
|
chr18_+_3247527
|
0.045
|
NM_006471
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr1_-_150552005
|
0.043
|
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr15_-_59665053
|
0.042
|
|
MYO1E
|
myosin IE
|
|
chr19_+_17666457
|
0.042
|
|
GLT25D1
|
glycosyltransferase 25 domain containing 1
|
|
chr7_-_5569200
|
0.040
|
|
ACTB
|
actin, beta
|
|
chr12_-_91505541
|
0.040
|
NM_002345
|
LUM
|
lumican
|
|
chr2_+_55459038
|
0.040
|
NM_001135592
|
RPS27A
|
ribosomal protein S27a
|
|
chr8_-_62602285
|
0.039
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr2_+_74120092
|
0.038
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr19_-_12792577
|
0.037
|
|
DHPS
|
deoxyhypusine synthase
|
|
chr9_-_79520878
|
0.035
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr3_+_187871473
|
0.034
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_+_114043413
|
0.034
|
NM_058222
|
TECTB
|
tectorin beta
|
|
chr1_+_151512760
|
0.034
|
NM_001126337
NM_020127
|
TUFT1
|
tuftelin 1
|
|
chr1_-_35324645
|
0.032
|
NM_001164824
|
C1orf212
|
chromosome 1 open reading frame 212
|
|
chr2_-_74607425
|
0.031
|
NM_001135040
NM_001190837
NM_004082
|
DCTN1
|
dynactin 1
|
|
chr18_-_65182507
|
0.031
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr17_+_72439187
|
0.030
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr1_+_50574601
|
0.030
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_+_154975041
|
0.030
|
NM_001252405
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr12_+_6881669
|
0.029
|
NM_002286
|
LAG3
|
lymphocyte-activation gene 3
|
|
chr14_-_53417808
|
0.029
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr3_-_9834065
|
0.028
|
|
TADA3
|
transcriptional adaptor 3
|
|
chr1_-_31845818
|
0.028
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr9_+_140772228
|
0.027
|
NM_000718
NM_001243812
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr1_+_19970674
|
0.027
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr6_-_26108320
|
0.026
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr6_+_158402916
|
0.026
|
|
SYNJ2
|
synaptojanin 2
|
|
chr12_+_54426831
|
0.026
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr21_+_35190586
|
0.025
|
|
ITSN1
|
intersectin 1 (SH3 domain protein)
|
|
chr2_-_216248184
|
0.025
|
|
FN1
|
fibronectin 1
|