|
chr6_+_26217145
|
4.713
|
NM_021052
|
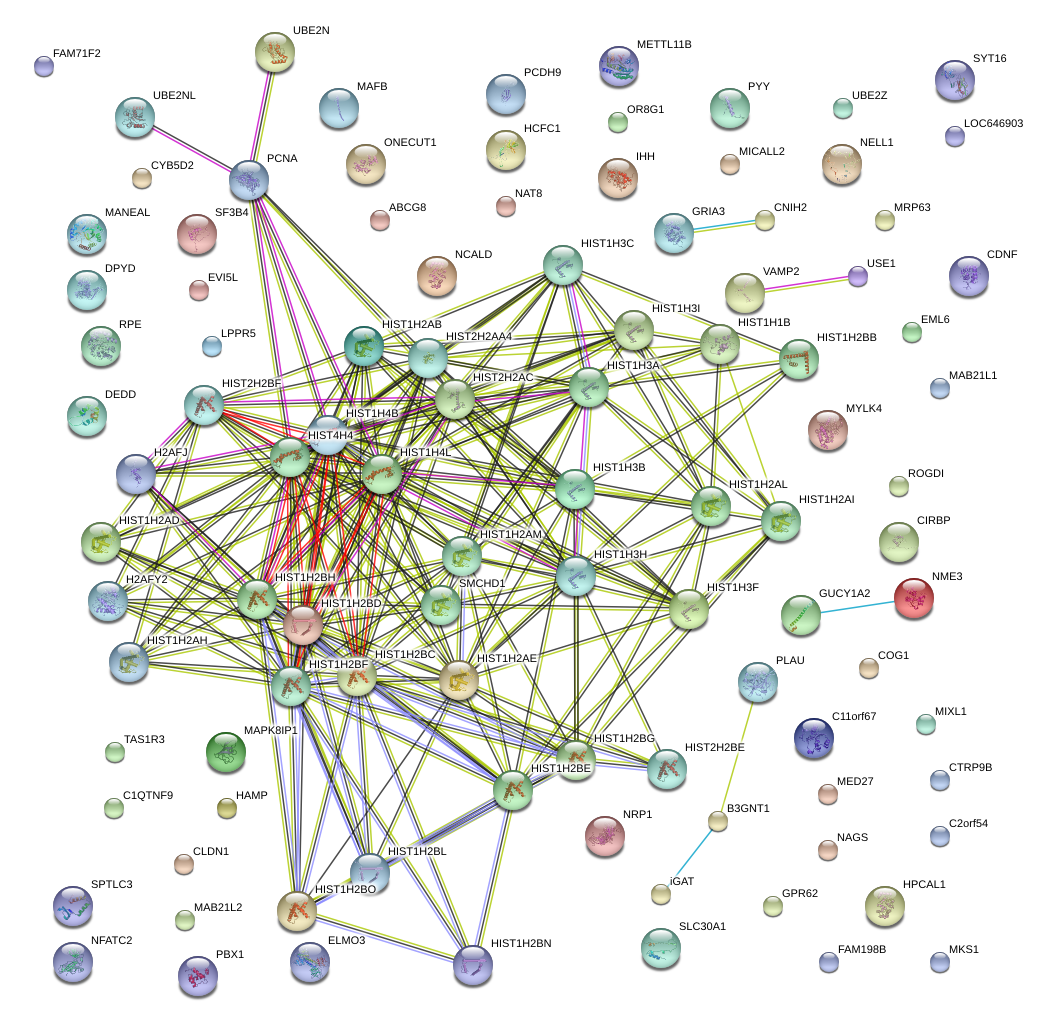
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr16_-_1821503
|
4.500
|
|
NME3
|
non-metastatic cells 3, protein expressed in
|
|
chr6_+_27775898
|
4.443
|
NM_003509
|
HIST1H2AI
HIST1H2AM
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H2am
histone cluster 1, H3f
|
|
chr6_+_26045611
|
3.843
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr1_-_99470177
|
3.827
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr1_-_211751719
|
3.506
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr6_-_27775683
|
3.397
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr6_-_26027479
|
3.160
|
NM_003544
|
HIST1H4B
|
histone cluster 1, H4b
|
|
chr6_-_26032287
|
2.963
|
NM_003537
|
HIST1H3B
|
histone cluster 1, H3b
|
|
chr6_+_27833033
|
2.654
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr1_-_211751953
|
2.584
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr6_-_26043884
|
2.369
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr6_+_26199712
|
2.335
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr20_-_44563573
|
2.252
|
|
FLJ40606
|
uncharacterized LOC643549
|
|
chr6_-_27840086
|
2.217
|
NM_003533
|
HIST1H3I
|
histone cluster 1, H3i
|
|
chr3_+_149689065
|
2.179
|
|
LOC646903
|
uncharacterized LOC646903
|
|
chr19_+_17326154
|
2.136
|
NM_018467
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae)
|
|
chr2_+_54952148
|
2.115
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr17_-_70588942
|
2.027
|
|
LOC100499467
|
uncharacterized LOC100499467
|
|
chr6_+_27114860
|
1.987
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chrX_+_142967172
|
1.975
|
NM_001012989
|
UBE2NL
|
ubiquitin-conjugating enzyme E2N-like
|
|
chr6_+_27861202
|
1.971
|
NM_003527
|
HIST1H2BO
|
histone cluster 1, H2bo
|
|
chr4_+_151503076
|
1.957
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr1_-_211752052
|
1.941
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr15_-_53082087
|
1.934
|
NM_004498
|
ONECUT1
|
one cut homeobox 1
|
|
chr7_-_1498961
|
1.917
|
|
MICALL2
|
MICAL-like 2
|
|
chr2_-_241835572
|
1.855
|
NM_001085437
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr19_+_35773409
|
1.775
|
NM_021175
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr19_+_17326196
|
1.644
|
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae)
|
|
chr6_-_27835308
|
1.621
|
NM_005322
|
HIST1H1B
|
histone cluster 1, H1b
|
|
chr7_-_1499107
|
1.618
|
NM_182924
|
MICALL2
|
MICAL-like 2
|
|
chr2_+_44066102
|
1.584
|
NM_022437
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8
|
|
chr19_+_1270926
|
1.509
|
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr10_+_71812536
|
1.495
|
|
H2AFY2
|
H2A histone family, member Y2
|
|
chrX_+_122318095
|
1.490
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr20_-_39317875
|
1.417
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr1_+_170115187
|
1.415
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr16_-_1821709
|
1.373
|
NM_002513
|
NME3
|
non-metastatic cells 3, protein expressed in
|
|
chr9_-_134955243
|
1.369
|
NM_004269
|
CRSP8P
MED27
|
mediator complex subunit 27 pseudogene
mediator complex subunit 27
|
|
chr10_-_33623299
|
1.367
|
|
NRP1
|
neuropilin 1
|
|
chr14_+_62462540
|
1.358
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr11_-_106888829
|
1.294
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr2_-_219925237
|
1.287
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr6_-_27860927
|
1.258
|
NM_003514
|
HIST1H2AM
|
histone cluster 1, H2am
|
|
chr11_+_124120422
|
1.249
|
NM_001002905
|
OR8G1
|
olfactory receptor, family 8, subfamily G, member 1
|
|
chr16_+_67233013
|
1.249
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr3_+_51989329
|
1.233
|
NM_080865
|
GPR62
|
G protein-coupled receptor 62
|
|
chr10_-_14879967
|
1.230
|
NM_001029954
|
CDNF
|
cerebral dopamine neurotrophic factor
|
|
chr1_+_38259458
|
1.223
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr4_-_159092509
|
1.217
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr1_-_98386458
|
1.186
|
|
DPYD
|
dihydropyrimidine dehydrogenase
|
|
chr1_-_149858114
|
1.160
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr13_-_67804432
|
1.155
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr4_-_146859960
|
1.154
|
|
|
|
|
chr16_-_4852571
|
1.150
|
NM_024589
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr11_+_45907198
|
1.149
|
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr6_-_26216871
|
1.137
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr18_-_53068942
|
1.132
|
|
|
|
|
chr20_-_50179167
|
1.096
|
NM_001136021
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chrX_-_153236621
|
1.095
|
NM_005334
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr19_+_7895110
|
1.094
|
NM_145245
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr17_-_8066254
|
1.078
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr20_+_12989616
|
1.075
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr2_-_73928466
|
1.073
|
NM_016347
|
NAT8B
|
N-acetyltransferase 8B (GCN5-related, putative, gene/pseudogene)
|
|
chr7_+_128312345
|
1.062
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr6_+_26183957
|
1.059
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr13_-_24471124
|
1.045
|
NM_001007537
|
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B
|
|
chr17_-_56296587
|
1.044
|
NM_017777
|
MKS1
|
Meckel syndrome, type 1
|
|
chr17_+_56065408
|
1.043
|
|
|
|
|
chr6_-_2751153
|
1.042
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr1_+_164528914
|
1.040
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr20_-_44516237
|
1.037
|
NM_080608
|
SPATA25
|
spermatogenesis associated 25
|
|
chr1_-_149783910
|
1.035
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr13_+_21750752
|
1.034
|
|
MRP63
|
mitochondrial ribosomal protein 63
|
|
chr10_+_105005643
|
1.019
|
NM_001143909
|
LOC729020
|
rcRPE
|
|
chr17_+_71189186
|
1.018
|
|
COG1
|
component of oligomeric golgi complex 1
|
|
chr1_+_38261079
|
1.013
|
NM_152496
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr1_-_98386594
|
1.013
|
NM_000110
NM_001160301
|
DPYD
|
dihydropyrimidine dehydrogenase
|
|
chr1_-_161102457
|
1.012
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr10_+_75673346
|
1.006
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr3_-_190040192
|
1.000
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr6_-_26033795
|
1.000
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr1_-_98386525
|
0.991
|
|
DPYD
|
dihydropyrimidine dehydrogenase
|
|
chr1_-_149900143
|
0.990
|
NM_005850
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa
|
|
chr11_-_66115016
|
0.984
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr17_-_42081836
|
0.977
|
NM_004160
|
PYY
|
peptide YY
|
|
chr11_+_77532159
|
0.977
|
NM_024684
|
C11orf67
|
chromosome 11 open reading frame 67
|
|
chr17_+_71189144
|
0.972
|
NM_018714
|
COG1
|
component of oligomeric golgi complex 1
|
|
chr1_+_1266722
|
0.965
|
NM_152228
|
TAS1R3
|
taste receptor, type 1, member 3
|
|
chr11_+_66045645
|
0.964
|
NM_182553
|
CNIH2
|
cornichon homolog 2 (Drosophila)
|
|
chr1_+_149858466
|
0.964
|
NM_003517
|
HIST2H2AC
|
histone cluster 2, H2ac
|
|
chr20_-_5100443
|
0.960
|
NM_182649
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr17_+_42081913
|
0.957
|
NM_153006
|
NAGS
|
N-acetylglutamate synthase
|
|
chr1_+_226411318
|
0.955
|
NM_031944
|
MIXL1
|
Mix paired-like homeobox
|
|
chr17_+_4046941
|
0.935
|
|
CYB5D2
|
cytochrome b5 domain containing 2
|
|
chr18_-_5892102
|
0.935
|
NM_001080209
|
TMEM200C
|
transmembrane protein 200C
|
|
chr2_+_63277936
|
0.922
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr6_+_27806322
|
0.916
|
NM_003520
|
HIST1H2BN
|
histone cluster 1, H2bn
|
|
chr15_-_75017710
|
0.908
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr1_+_51435911
|
0.907
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr9_-_130829598
|
0.895
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr17_+_15848422
|
0.890
|
|
ADORA2B
|
adenosine A2b receptor
|
|
chr11_+_111473099
|
0.886
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr12_-_48298765
|
0.886
|
NM_000376
NM_001017535
NM_001017536
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr5_-_139943829
|
0.885
|
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr15_-_72612247
|
0.879
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr14_-_94844881
|
0.876
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr15_-_52587851
|
0.875
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr19_+_797452
|
0.873
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr6_-_134497069
|
0.872
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr15_-_41408409
|
0.871
|
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr7_+_44836309
|
0.869
|
|
PPIA
|
peptidylprolyl isomerase A (cyclophilin A)
|
|
chr7_-_140624280
|
0.869
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr2_-_74734498
|
0.869
|
|
PCGF1
|
polycomb group ring finger 1
|
|
chr7_-_139477437
|
0.863
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr2_+_233734993
|
0.862
|
NM_206895
|
C2orf82
|
chromosome 2 open reading frame 82
|
|
chr7_+_114055048
|
0.853
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr11_-_2182408
|
0.848
|
NM_000207
NM_001185097
NM_001185098
NM_001042376
|
INS
INS-IGF2
|
insulin
INS-IGF2 readthrough
|
|
chr19_-_41870001
|
0.842
|
NM_030578
|
B9D2
|
B9 protein domain 2
|
|
chr7_+_114055234
|
0.840
|
|
FOXP2
|
forkhead box P2
|
|
chr1_+_110026452
|
0.840
|
NM_153340
|
ATXN7L2
|
ataxin 7-like 2
|
|
chr10_+_71812356
|
0.840
|
NM_018649
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr3_-_9290592
|
0.836
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr20_+_44563310
|
0.832
|
NM_022104
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1
|
|
chr1_-_161102367
|
0.829
|
|
DEDD
|
death effector domain containing
|
|
chr19_+_7911385
|
0.828
|
NM_001159944
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr17_+_42634598
|
0.821
|
NM_001466
|
FZD2
|
frizzled family receptor 2
|
|
chr18_-_47814024
|
0.820
|
|
CXXC1
|
CXXC finger protein 1
|
|
chr19_+_797448
|
0.820
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chrX_+_153059903
|
0.814
|
NM_001204526
NM_006280
|
SSR4
|
signal sequence receptor, delta
|
|
chr11_-_116693907
|
0.807
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr7_+_39017608
|
0.807
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr1_+_145507556
|
0.805
|
NM_005105
|
RBM8A
|
RNA binding motif protein 8A
|
|
chr15_+_42131010
|
0.803
|
NM_001114633
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic)
|
|
chr1_+_161719540
|
0.800
|
NM_007240
|
DUSP12
|
dual specificity phosphatase 12
|
|
chr1_+_97187174
|
0.796
|
NM_021190
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr2_+_189839132
|
0.796
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_-_15114495
|
0.793
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr9_-_34522972
|
0.792
|
NM_198573
|
ENHO
|
energy homeostasis associated
|
|
chr17_+_78388944
|
0.791
|
NM_001164637
NM_001164638
NM_173627
|
ENDOV
|
endonuclease V
|
|
chr3_-_59035700
|
0.790
|
NM_198463
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr22_-_36236264
|
0.788
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr2_+_203776930
|
0.788
|
NM_001104586
NM_024744
|
ALS2CR8
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 8
|
|
chr3_-_133748828
|
0.787
|
NM_005630
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chrX_+_106171328
|
0.787
|
|
CLDN2
|
claudin 2
|
|
chr16_-_29625037
|
0.785
|
|
|
|
|
chr6_-_88854989
|
0.785
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr6_-_26189303
|
0.781
|
NM_003539
|
HIST1H4D
|
histone cluster 1, H4d
|
|
chr1_+_180897572
|
0.778
|
|
|
|
|
chr20_-_5107185
|
0.778
|
NM_002592
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr19_+_797390
|
0.778
|
NM_002819
NM_031990
NM_031991
NM_175847
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr11_-_66103874
|
0.777
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr11_+_1331048
|
0.775
|
|
LOC255512
|
uncharacterized LOC255512
|
|
chr13_+_93879011
|
0.774
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr5_-_27038650
|
0.773
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr5_+_41925355
|
0.773
|
NM_012176
NM_033484
|
FBXO4
|
F-box protein 4
|
|
chr12_-_102591547
|
0.771
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr9_-_134955056
|
0.771
|
|
MED27
|
mediator complex subunit 27
|
|
chr7_+_114055171
|
0.769
|
|
FOXP2
|
forkhead box P2
|
|
chr18_-_47814005
|
0.767
|
|
CXXC1
|
CXXC finger protein 1
|
|
chr6_-_134496833
|
0.767
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_+_106546736
|
0.765
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr16_+_56642477
|
0.764
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr13_-_37679800
|
0.764
|
NM_145203
|
CSNK1A1L
|
casein kinase 1, alpha 1-like
|
|
chr1_-_149899623
|
0.764
|
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa
|
|
chr2_+_39893021
|
0.762
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr12_-_6665220
|
0.760
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr1_-_153606567
|
0.755
|
NM_001024210
|
S100A13
|
S100 calcium binding protein A13
|
|
chr17_+_78010430
|
0.753
|
NM_001243342
NM_017950
|
CCDC40
|
coiled-coil domain containing 40
|
|
chr5_+_167956545
|
0.752
|
|
|
|
|
chr6_+_41020939
|
0.752
|
NM_006789
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2
|
|
chr2_+_155555092
|
0.751
|
NM_002239
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3
|
|
chr16_-_30621628
|
0.746
|
NM_138447
|
ZNF689
|
zinc finger protein 689
|
|
chr9_+_131902208
|
0.745
|
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4
|
|
chr7_-_16505292
|
0.745
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr20_+_10199455
|
0.744
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr6_+_75994713
|
0.741
|
|
LOC100506804
|
uncharacterized LOC100506804
|
|
chr1_+_27648622
|
0.740
|
NM_032125
|
TMEM222
|
transmembrane protein 222
|
|
chr19_-_38746978
|
0.739
|
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr9_+_130026755
|
0.739
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr12_-_22697439
|
0.738
|
NM_014802
|
KIAA0528
|
KIAA0528
|
|
chr20_-_44600939
|
0.734
|
|
ZNF335
|
zinc finger protein 335
|
|
chr11_+_111749947
|
0.733
|
NM_022761
|
C11orf1
|
chromosome 11 open reading frame 1
|
|
chr10_-_90342947
|
0.732
|
NM_001031709
NM_018363
|
RNLS
|
renalase, FAD-dependent amine oxidase
|
|
chr2_-_190044330
|
0.732
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr19_+_797444
|
0.731
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr4_+_75858205
|
0.731
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr1_+_61547625
|
0.729
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr11_-_115375065
|
0.729
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr7_-_73184535
|
0.729
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr9_+_214816
|
0.727
|
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr19_+_1269244
|
0.725
|
NM_001280
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr17_+_47865980
|
0.725
|
NM_001199155
NM_001199156
NM_001199157
NM_001199158
NM_007067
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr1_-_220101781
|
0.725
|
|
|
|
|
chr5_+_180650709
|
0.724
|
|
TRIM41
|
tripartite motif containing 41
|
|
chr1_+_27023932
|
0.723
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr11_-_117186364
|
0.722
|
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr22_-_33402648
|
0.721
|
NM_003490
NM_133633
|
SYN3
|
synapsin III
|
|
chr9_-_140115774
|
0.719
|
NM_031297
|
RNF208
|
ring finger protein 208
|
|
chr11_-_116968986
|
0.715
|
NM_025164
|
SIK3
|
SIK family kinase 3
|