MYCN
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| MYCN | NMYC, ODED, MODED, N-myc, bHLHe37,bHLHe37, MYCNOT | v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog | NM_005378 |  |
 |
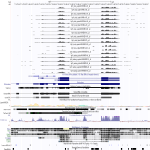 |
 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
This gene is a member of the MYC family and encodes a protein with a basic helix-loop-helix (bHLH) domain. This protein is located in the nucleus and must dimerize with another bHLH protein in order to bind DNA. Amplification of this gene is associated with a variety of tumors, most notably neuroblastomas. Multiple alternatively spliced transcript variants encoding different isoforms have been found for this gene.
The transcriptional regulator MYCN has to dimerize, preferentially with the Max protein, to bind to DNA by a basic helix-loop-helix motif [Ma et al., Oncogene, 1993, 8:1093-1098]. N-myc has been shown to both up- and down-regulate many target genes, in addition, this factor modulates the expression of several classes of non-coding RNAs, in particular microRNAs [Buechner and Einvik, Mol Cancer Res, 2012, 10:1243-1253]. N-myc mRNA was strongly down-regulated (20X) in human cells infected with EBOV, whereas in MARV-infected cells, only slight down-regulation was observed (3X). This gene is not part of the bat transcriptome assembly. Due to the very strong down-regulation of n-myc, it seems to be very important for an EBOV infection to deplete this factor from infected cells.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 238 | 275 | 373 | 195 | 340 | 15 | 218 | 322 | 102 | |
Read_Max | R. aegyptiacus | Transcriptome | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | |
Read_Max | R. aegyptiacus | Genome | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | |
DESeq | H. sapiens | Genome | 2852.55 | 3707.22 | 4918.46 | 2654.23 | 3531.65 | 69.43 | 2369.45 | 3366.27 | 1338.44 | |
DESeq | R. aegyptiacus | Transcriptome | 1.0 | 1.0 | 2.22 | 1.0 | 1.0 | 1.0 | 3.22 | 1.0 | 2.75 | |
DESeq | R. aegyptiacus | Genome | 2.58 | 0.0 | 1.12 | 0.99 | 0.0 | 0.0 | 2.25 | 0.0 | 0.0 |