DUSP8
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| DUSP8 | C11orf81, DUS8_HUMAN, NM_004420, NP_004411, Q13202, Q86SS8, VH5,HVH-5, HB5, FLJ42958 | dual specificity phosphatase 8 | NM_004420 | 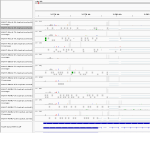 |
 |
 |
 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
The protein encoded by this gene is a member of the dual specificity protein phosphatase subfamily. These phosphatases inactivate their target kinases by dephosphorylating both the phosphoserine/threonine and phosphotyrosine residues. They negatively regulate members of the mitogen-activated protein (MAP) kinase superfamily (MAPK/ERK, SAPK/JNK, p38), which is associated with cellular proliferation and differentiation. Different members of the family of dual specificity phosphatases show distinct substrate specificities for various MAP kinases, different tissue distribution and subcellular localization, and different modes of inducibility of their expression by extracellular stimuli. This gene product inactivates SAPK/JNK and p38, is expressed predominantly in the adult brain, heart, and skeletal muscle, is localized in the cytoplasm, and is induced by nerve growth factor and insulin. An intronless pseudogene for DUSP8 is present on chromosome 10q11.2
An interesting gene. Seems to be not differentially expressed in bat cells, but has a very high foldchange in Ebola infected cells in human cells after 23 h.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 31 | 25 | 13 | 30 | 26 | 626 | 44 | 29 | 26 | |
Read_Max | R. aegyptiacus | Transcriptome | 48 | 53 | 35 | 48 | 42 | 72 | 24 | 42 | 58 | |
Read_Max | R. aegyptiacus | Genome | 54 | 43 | 26 | 37 | 27 | 38 | 23 | 57 | 69 | |
DESeq | H. sapiens | Genome | 429.24 | 298.8 | 223.39 | 396.82 | 282.28 | 9033.49 | 572.94 | 427.85 | 538.15 | |
DESeq | R. aegyptiacus | Transcriptome | 184.99 | 246.98 | 291.73 | 229.0 | 244.11 | 474.49 | 167.86 | 263.73 | 368.38 | |
DESeq | R. aegyptiacus | Genome | 332.58 | 451.78 | 548.96 | 386.11 | 394.0 | 757.14 | 281.13 | 454.36 | 595.05 |