PLAU
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| PLAU | ATF, QPD, UPA, URK, u-PA, BDPLT5,URK | plasminogen activator, urokinase | NM_002658 |  |
 |
 |
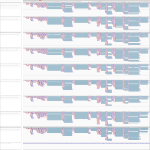 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
This gene encodes a serine protease involved in degradation of the extracellular matrix and possibly tumor cell migration and proliferation. A specific polymorphism in this gene may be associated with late-onset Alzheimer's disease and also with decreased affinity for fibrin-binding. This protein converts plasminogen to plasmin by specific cleavage of an Arg-Val bond in plasminogen. Plasmin in turn cleaves this protein at a Lys-Ile bond to form a two-chain derivative in which a single disulfide bond connects the amino-terminal A-chain to the catalytically active, carboxy-terminal B-chain. This two-chain derivative is also called HMW-uPA (high molecular weight uPA). HMW-uPA can be further processed into LMW-uPA (low molecular weight uPA) by cleavage of chain A into a short chain A (A1) and an amino-terminal fragment. LMW-uPA is proteolytically active but does not bind to the uPA receptor. Alternatively spliced transcript variants encoding different isoforms have been found for this gene.
Low, but clear expression in human cells and down-regulated in Ebola- and Marburg-infected cells after 23 h. In the bat cells homolog, the expression is about 10-fold higher and also up-regulated in Ebola and Marburg-infected cells after 23 h.
Antisense to C10orf55 (see Section~\refapg:hg19.apg.00166).
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 14 | 33 | 11 | 19 | 29 | 6 | 23 | 23 | 2 | |
Read_Max | R. aegyptiacus | Transcriptome | 199 | 198 | 216 | 211 | 164 | 366 | 158 | 241 | 479 | |
Read_Max | R. aegyptiacus | Genome | 208 | 209 | 173 | 195 | 136 | 194 | 129 | 280 | 484 | |
DESeq | H. sapiens | Genome | 128.86 | 277.39 | 123.12 | 208.6 | 183.91 | 16.79 | 198.37 | 184.05 | 8.61 | |
DESeq | R. aegyptiacus | Transcriptome | 1285.57 | 1449.67 | 1899.27 | 1544.82 | 1526.46 | 2555.81 | 1305.85 | 1524.53 | 2753.71 | |
DESeq | R. aegyptiacus | Genome | 1612.22 | 1835.41 | 2250.52 | 1896.96 | 1937.61 | 3216.53 | 1663.18 | 1864.22 | 3465.01 |