HIST1H3C
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| HIST1H3C | HIST1H3C, H3.1, H3/c, H3FC,H3/c | histone cluster 1, H3c | NM_003531 |  |
 |
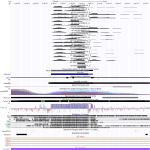 |
NONE |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Two molecules of each of the four core histones (H2A, H2B, H3, and H4) form an octamer, around which approximately 146 bp of DNA is wrapped in repeating units, called nucleosomes. The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene is intronless and encodes a member of the histone H3 family. Transcripts from this gene lack polyA tails but instead contain a palindromic termination element.
Manual screening of HIST1H3C transcriptome data did not reveal any remarkable alteration due to Ebola or Marburg virus infection.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 12 | 8 | 5 | 9 | 11 | 11 | 14 | 9 | 1 | |
Read_Max | R. aegyptiacus | Transcriptome | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
Read_Max | R. aegyptiacus | Genome | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
DESeq | H. sapiens | Genome | 31.44 | 27.52 | 10.93 | 27.24 | 18.07 | 28.63 | 29.95 | 21.78 | 2.09 | |
DESeq | R. aegyptiacus | Transcriptome | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
DESeq | R. aegyptiacus | Genome | NA | NA | NA | NA | NA | NA | NA | NA | NA |