ACADSB
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| ACADSB | SBCAD, ACAD7 | acyl-CoA dehydrogenase, short/branched chain | NM_001609 |  |
 |
 |
NONE |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
Short/branched chain acyl-CoA dehydrogenase(ACADSB) is a member of the acyl-CoA dehydrogenase family of enzymes that catalyze the dehydrogenation of acyl-CoA derivatives in the metabolism of fatty acids or branch chained amino acids. Substrate specificity is the primary characteristic used to define members of this gene family. The ACADSB gene product has the greatest activity towards the short branched chain acyl-CoA derivative, (S)-2-methylbutyryl-CoA, but also reacts significantly with other 2-methyl branched chain substrates and with short straight chain acyl-CoAs. The cDNA encodes for a mitochondrial precursor protein which is cleaved upon mitochondrial import and predicted to yield a mature peptide of approximately 43.7-KDa. [provided by RefSeq, Jul 2008]. Sequence Note: The 3' UTR extension represented by the RefSeq transcript record was derived from genomic sequence data to optimize consistency to the reference genome assembly. The extent of the UTR extension and the location of the polyA site was based on transcript alignments.
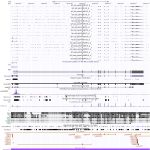
The gene, whose homolog was not expressed in bat cells, shows in human cells for Ebola and Marburg infection a huge upregulation for the 7 h sample and is downregulated afterwards, for Ebola nearly turned off. All samples show a slight expression of the first intron with length of nearly 25\,000\,nt.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 704 | 794 | 719 | 893 | 1121 | 86 | 904 | 1165 | 371 | |
Read_Max | R. aegyptiacus | Transcriptome | 2 | NA | 4 | 5 | NA | 2 | 1 | 2 | 1 | |
Read_Max | R. aegyptiacus | Genome | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
DESeq | H. sapiens | Genome | 10839.94 | 10813.69 | 10566.64 | 12118.48 | 12306.64 | 1332.71 | 11319.57 | 10210.3 | 5965.1 | |
DESeq | R. aegyptiacus | Transcriptome | 4.36 | 1.0 | 10.77 | 6.49 | 1.0 | 4.85 | 3.22 | 2.65 | 1.87 | |
DESeq | R. aegyptiacus | Genome | NA | NA | NA | NA | NA | NA | NA | NA | NA |