MAP1B
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| MAP1B | FUTSCH, MAP5, PPP1R102,MAP5 | microtubule-associated protein 1B | NM_005909 |  |
 |
 |
 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
microtubule-associated protein 1B is involved in axon extension, negative regulation of intracellular transport and dendrite development. Facilitates tyrosination of alpha-tubulin in neuronal microtubules (By similarity). Phosphorylated MAP1B may play a role in the cytoskeletal changes that accompany neurite extension. Possibly MAP1B binds to at least two tubulin subunits in the polymer, and this bridging of subunits might be involved in nucleating microtubule polymerization and in stabilizing microtubules. Acts as a positive cofactor in DAPK1-mediated autophagic vesicle formation and membrane blebbing.
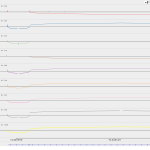
It does not present a particularly interesting differential expression pattern in the samples analyzed, except for the slight upregulation after 3 hours of infection with Ebola in humans and the slight downregulation after 23 hours of infection with Marburg virus in humans.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 18 | 24 | 25 | 17 | 46 | 50 | 22 | 30 | 100 | |
Read_Max | R. aegyptiacus | Transcriptome | 476 | 282 | 462 | 730 | 646 | 437 | 524 | 691 | 590 | |
Read_Max | R. aegyptiacus | Genome | 512 | 679 | 364 | 266 | 495 | 242 | 430 | 791 | 635 | |
DESeq | H. sapiens | Genome | 655.54 | 764.88 | 670.16 | 555.37 | 824.52 | 977.41 | 550.13 | 812.34 | 4005.7 | |
DESeq | R. aegyptiacus | Transcriptome | 10394.38 | 7842.53 | 10073.79 | 11712.4 | 12639.43 | 9744.2 | 11850.35 | 9480.92 | 11516.54 | |
DESeq | R. aegyptiacus | Genome | 11563.09 | 12533.9 | 10140.19 | 9305.06 | 14088.96 | 10936.72 | 13522.5 | 10451.03 | 12470.43 |