TRIM32
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| TRIM32 | HT2A, NM_001099679, NP_036342, Q13049, Q9NQP8, TRI32_HUMAN,HT2A, TATIP, BBS11 | tripartite motif containing 32 | NM_012210 |  |
 |
 |
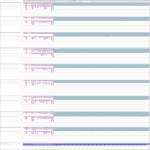 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. The protein localizes to cytoplasmic bodies. The protein has also been localized to the nucleus, where it interacts with the activation domain of the HIV-1 Tat protein. The Tat protein activates transcription of HIV-1 genes.
While for MOCK and EBOV cells a downregulation was observed between 7 h and 23 h p.i., there was an upregulation for MARV infected cells. No significant differential expression was seen within the bat.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 63 | 54 | 68 | 65 | 64 | 24 | 71 | 73 | 52 | |
Read_Max | R. aegyptiacus | Transcriptome | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
Read_Max | R. aegyptiacus | Genome | 149 | 127 | 107 | 135 | 105 | 57 | 105 | 185 | 131 | |
DESeq | H. sapiens | Genome | 871.69 | 869.93 | 867.74 | 934.08 | 679.0 | 191.81 | 901.01 | 793.16 | 840.43 | |
DESeq | R. aegyptiacus | Transcriptome | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
DESeq | R. aegyptiacus | Genome | 1542.61 | 1391.84 | 1458.32 | 1718.22 | 1564.82 | 1361.54 | 1839.73 | 1455.85 | 1324.53 |