MCM4
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| MCM4 | AK309812,CDC54, hCdc21, P1-Cdc21, MGC33310 | minichromosome maintenance complex component 4 | NM_005914 |  |
 |
 |
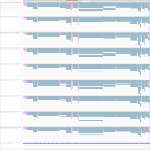 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
The protein encoded by the gene MCM4 is one of the highly conserved mini-chromosome maintenance proteins (MCM) that are essential for the initiation of eukaryotic genome replication. The hexameric protein complex formed by MCM proteins is a key component of the pre-replication complex (pre\_RC) and may be involved in the formation of replication forks and in the recruitment of other DNA replication related proteins. The MCM complex consisting of this protein and MCM2, 6 and 7 proteins possesses DNA helicase activity, and may act as a DNA unwinding enzyme. The phosphorylation of this protein by CDC2 kinase reduces the DNA helicase activity and chromatin binding of the MCM complex. This gene is mapped to a region on the chromosome 8 head-to-head next to the PRKDC/DNA-PK, a DNA-activated protein kinase involved in the repair of DNA double-strand breaks. Alternatively spliced transcript variants encoding the same protein have been reported.
For this gene we observed a downregulation after 23 h for all probes, which was most significant within the cells infected with the Marburg virus (3x). However this gene was also less expressed at this timepoint in the bat cells for all probes. Interestingly, there were splice variants, which were unique for the Marburg infected cells (exon 7-10) and Ebola infected cells (exon 6-9) after 7 h.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 919 | 884 | 650 | 853 | 856 | 231 | 1022 | 1102 | 331 | |
Read_Max | R. aegyptiacus | Transcriptome | 1687 | 1500 | 690 | 1396 | 1245 | 946 | 1237 | 1592 | 1128 | |
Read_Max | R. aegyptiacus | Genome | 1612 | 1301 | 519 | 1294 | 926 | 507 | 949 | 1738 | 1113 | |
DESeq | H. sapiens | Genome | 11216.42 | 9984.54 | 9847.83 | 11055.36 | 9654.79 | 2936.82 | 11075.71 | 9570.02 | 5821.57 | |
DESeq | R. aegyptiacus | Transcriptome | 11270.64 | 11439.71 | 5815.51 | 9768.43 | 9890.76 | 6964.83 | 11826.99 | 9824.62 | 6871.84 | |
DESeq | R. aegyptiacus | Genome | 13640.24 | 11740.78 | 6386.71 | 14475.52 | 11684.81 | 8494.3 | 14313.04 | 11511.2 | 7893.23 |