TRIM2
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| TRIM2 | KIAA0517, NM_001130067, NP_001123539, O60272, Q9BSI9, Q9C040, Q9UFZ1, RNF86, TRIM2_HUMAN | tripartite motif containing 2 |  |
 |
 |
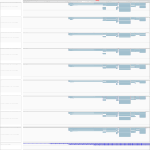 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. The protein localizes to cytoplasmic filaments. It plays a neuroprotective role and functions as an E3-ubiquitin ligase in proteasome-mediated degradation of target proteins. Mutations in this gene can cause early-onset axonal neuropathy. Alternative splicing results in multiple transcript variants.
For this gene we saw differential expression within the human cells. For the infected cells first an upregulation (between 3 h and 7 h p.i.) and then a downregulation (between 7 h and 23 h p.i.) took place.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 132 | 174 | 152 | 144 | 191 | 175 | 152 | 215 | 250 | |
Read_Max | R. aegyptiacus | Transcriptome | 149 | 106 | 67 | 103 | 78 | 92 | 122 | 126 | 139 | |
Read_Max | R. aegyptiacus | Genome | 154 | 106 | 57 | 98 | 86 | 56 | 103 | 156 | 162 | |
DESeq | H. sapiens | Genome | 3063.63 | 3745.97 | 4198.66 | 2842.44 | 3939.75 | 1182.7 | 2622.97 | 3368.66 | 3715.38 | |
DESeq | R. aegyptiacus | Transcriptome | 1328.42 | 996.74 | 1111.38 | 988.09 | 1077.47 | 935.16 | 1265.81 | 1058.55 | 1272.83 | |
DESeq | R. aegyptiacus | Genome | 2932.24 | 2556.43 | 2331.98 | 2442.05 | 2751.27 | 2314.49 | 3135.2 | 2645.27 | 2990.68 |