MAP4K2
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| MAP4K2 | MAP4K2, BL44, GCK, RAB8IP,GCK | mitogen-activated protein kinase kinase kinase kinase 2 | NM_004579 |  |
 |
 |
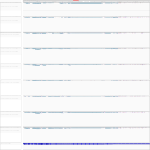 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
Serine/threonine-protein kinase which acts as an essential component of the MAP kinase signal transduction pathway. Acts as a MAPK kinase kinase kinase (MAP4K) and is an upstream activator of the stress-activated protein kinase/c-Jun N-terminal kinase (SAP/JNK) signaling pathway and to a lesser extend of the p38 MAPKs signaling pathway. Required for the efficient activation of JNKs by TRAF6-dependent stimuli, including pathogen-associated molecular patterns (PAMPs) such as polyinosine-polycytidine (poly(IC)), lipopolysaccharides (LPS), lipid A, peptidoglycan (PGN), or bacterial flagellin. To a lesser degree, IL-1 and engagement of CD40 also stimulate MAP4K2-mediated JNKs activation. The requirement for MAP4K2/GCK is most pronounced for LPS signaling, and extends to LPS stimulation of c-Jun phosphorylation and induction of IL-8. Enhances MAP3K1 oligomerization, which may relieve N-terminal mediated MAP3K1 autoinhibition and lead to activation following autophosphorylation. Mediates also the SAP/JNK signaling pathway and the p38 MAPKs signaling pathway through activation of the MAP3Ks MAP3K10/MLK2 and MAP3K11/MLK3. May play a role in the regulation of vesicle targeting or fusion. regulation of vesicle targeting or fusion.
Its expression seems to be mostly unaffected by Ebola or Marburg virus infection. Some low abundant exons seem to be consistently excluded in the isoform formation due to Ebola virus infection, in contrast to mock treated or Marburg virus infected samples.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 17 | 24 | 14 | 14 | 18 | 16 | 17 | 18 | 17 | |
Read_Max | R. aegyptiacus | Transcriptome | 29 | 39 | 26 | 31 | 34 | 25 | 18 | 50 | 39 | |
Read_Max | R. aegyptiacus | Genome | 33 | 30 | 24 | 32 | 31 | 15 | 14 | 53 | 43 | |
DESeq | H. sapiens | Genome | 134.95 | 184.58 | 150.92 | 147.01 | 143.27 | 114.17 | 135.21 | 170.46 | 158.67 | |
DESeq | R. aegyptiacus | Transcriptome | 290.01 | 316.88 | 321.04 | 272.95 | 266.01 | 216.57 | 231.27 | 347.18 | 329.02 | |
DESeq | R. aegyptiacus | Genome | 537.12 | 522.06 | 617.03 | 534.23 | 531.28 | 382.49 | 413.83 | 609.78 | 512.0 |