IGF1R
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| IGF1R | B1B5Y2, EU826611, IGF1R_HUMAN, NM_000875, NP_000866, P08069, Q14CV2, Q9UCC0,JTK13, CD221, IGFIR, MGC18216, IGFR | insulin-like growth factor 1 receptor | NM_000875 |  |
 |
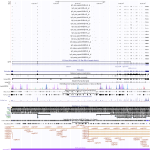 |
NONE |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
Homo sapiens insulin-like growth factor 1 receptor (IGF1R) receptor binds insulin-like growth factor with a high affinity. It has tyrosine kinase activity. The insulin-like growth factor I receptor plays a critical role in transformation events. Cleavage of the precursor generates alpha and beta subunits. It is highly overexpressed in most malignant tissues where it functions as an anti-apoptotic agent by enhancing cell survival. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene.
Receptor tyrosine kinase which mediates actions of insulin-like growth factor 1 (IGF1). Binds IGF1 with high affinity and IGF2 and insulin (INS) with a lower affinity. The activated IGF1R is involved in cell growth and survival control. IGF1R is crucial for tumor transformation and survival of malignant cell. Ligand binding activates the receptor kinase, leading to receptor autophosphorylation, and tyrosines phosphorylation of multiple substrates, that function as signaling adapter proteins including, the insulin-receptor substrates (IRS1/2), Shc and 14-3-3 proteins. Phosphorylation of IRSs proteins lead to the activation of two main signaling pathways: the PI3K-AKT/PKB pathway and the Ras-MAPK pathway. The result of activating the MAPK pathway is increased cellular proliferation, whereas activating the PI3K pathway inhibits apoptosis and stimulates protein synthesis. Phosphorylated IRS1 can activate the 85 kDa regulatory subunit of PI3K (PIK3R1), leading to activation of several downstream substrates, including protein AKT/PKB. AKT phosphorylation, in turn, enhances protein synthesis through mTOR activation and triggers the antiapoptotic effects of IGFIR through phosphorylation and inactivation of BAD. In parallel to PI3K-driven signaling, recruitment of Grb2/SOS by phosphorylated IRS1 or Shc leads to recruitment of Ras and activation of the ras-MAPK pathway. In addition to these two main signaling pathways IGF1R signals also through the Janus kinase/signal transducer and activator of transcription pathway (JAK/STAT). Phosphorylation of JAK proteins can lead to phosphorylation/activation of signal transducers and activators of transcription (STAT) proteins. In particular activation of STAT3, may be essential for the transforming activity of IGF1R. The JAK/STAT pathway activates gene transcription and may be responsible for the transforming activity. JNK kinases can also be activated by the IGF1R. IGF1 exerts inhibiting activities on JNK activation via phosphorylation and inhibition of MAP3K5/ASK1, which is able to directly associate with the IGF1R. When present in a hybrid receptor with INSR, binds IGF1. PubMed:12138094 shows that hybrid receptors composed of IGF1R and INSR isoform Long are activated with a high affinity by IGF1, with low affinity by IGF2 and not significantly activated by insulin, and that hybrid receptors composed of IGF1R and INSR isoform Short are activated by IGF1, IGF2 and insulin. In contrast, PubMed:16831875 shows that hybrid receptors composed of IGF1R and INSR isoform Long and hybrid receptors composed of IGF1R and INSR isoform Short have similar binding characteristics, both bind IGF1 and have a low affinity for insulin.
IGFR1 is very low expressed in HG19, but seems to be upregulated in the ebola infected samples. In contrast this gene is very strong expressed in RAE by decreasing amount of transcripts comparing MOCK with ebola and ebola with marburg infected samples. Downregulation can be observed for MOCK and ebola infection after 7 and 23h, while marburg is upregulated after 7h.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 20 | 21 | 19 | 21 | 26 | 44 | 23 | 24 | 29 | |
Read_Max | R. aegyptiacus | Transcriptome | 1019 | 901 | 629 | 799 | 609 | 657 | 654 | 664 | 797 | |
Read_Max | R. aegyptiacus | Genome | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
DESeq | H. sapiens | Genome | 750.93 | 668.0 | 595.7 | 608.98 | 657.86 | 1405.09 | 583.46 | 627.69 | 1287.33 | |
DESeq | R. aegyptiacus | Transcriptome | 20484.46 | 18796.53 | 18076.08 | 16776.96 | 16259.8 | 15509.53 | 17439.08 | 12748.57 | 14710.1 | |
DESeq | R. aegyptiacus | Genome | NA | NA | NA | NA | NA | NA | NA | NA | NA |