NRAS
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| NRAS | N-ras | neuroblastoma RAS viral (v-ras) oncogene homolog | NM_002524 |  |
 |
 |
 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
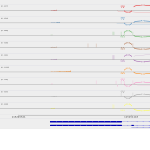
This is an N-ras oncogene encoding a membrane protein that shuttles betweenthe Golgi apparatus and the plasma membrane. This shuttling is regulatedthrough palmitoylation and depalmitoylation by the ZDHHC9-GOLGA7 complex.The encoded protein, which has intrinsic GTPase activity, is activated by aguanine nucleotide-exchange factor and inactivated by a GTPase activatingprotein. Mutations in this gene have been associated with somatic rectalcancer, follicular thyroid cancer, autoimmune lymphoproliferative syndrome,Noonan syndrome, and juvenile myelomonocytic leukemia.
It is expressed constantly in human cells as well as in bat cells but at a distincthigher level. %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
\igvfigure{\beginfigure[H]\centering\includegraphics[width=\textwidth]../../input/holy_folders/hg19.goi.00621/snapshots//hg19.goi.00621_igv.eps %\caption\igv of gene NRAS. % <- say something about the figure here\endfigure}
\sashimifigure{\beginfigure[H]\centering\includegraphics[width=\textwidth]../../input/holy_folders/hg19.goi.00621/snapshots//hg19.goi.00621_sashimi.eps %\caption\sashimi of gene NRAS. % <- say something about the figure here\endfigure}
\ucscfigure{\beginfigure[H]\centering\includegraphics[width=\textwidth]../../input/holy_folders/hg19.goi.00621/snapshots//hg19.goi.00621_ucsc.eps %\caption\ucsc of gene NRAS. % <- say something about the figure here\endfigure}
\setcounterfigure0\enddocument
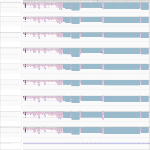
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 649 | 646 | 635 | 606 | 772 | 440 | 731 | 734 | 667 | |
Read_Max | R. aegyptiacus | Transcriptome | 682 | 581 | 367 | 567 | 454 | 453 | 456 | 604 | 510 | |
Read_Max | R. aegyptiacus | Genome | 761 | 574 | 348 | 580 | 400 | 286 | 420 | 762 | 628 | |
DESeq | H. sapiens | Genome | 10785.14 | 9184.96 | 10721.52 | 9832.55 | 10141.75 | 6959.6 | 9803.76 | 8165.57 | 12867.58 | |
DESeq | R. aegyptiacus | Transcriptome | 1751.85 | 1587.64 | 1449.74 | 1692.24 | 1661.16 | 1409.93 | 1725.23 | 1659.2 | 1494.13 | |
DESeq | R. aegyptiacus | Genome | 8506.24 | 8584.71 | 7230.24 | 7830.74 | 8641.11 | 8440.78 | 9549.53 | 8807.25 | 7867.55 |