MAP3K8
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| MAP3K8 | A8K2Q5, COT, D3DRX1, ESTF, M3K8_HUMAN, NM_005204, NP_005195, P41279, Q14275, Q5T855, Q9HC81, TPL2,Tpl-2, EST, c-COT, MEKK8 | mitogen-activated protein kinase kinase kinase 8 | NM_005204 | 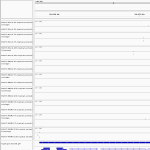 |
 |
 |
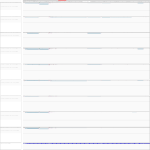 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
Homo sapiens mitogen-activated protein kinase kinase kinase 8 (MAP3K8), transcript variant 1 gen is an oncogene that encodes a member of the serine/threonine protein kinase family. The encoded protein localizes to the cytoplasm and can activate both the MAP kinase and JNK kinase pathways. This protein was shown to activate IkappaB kinases, and thus induce the nuclear production of NF-kappaB. This protein was also found to promote the production of TNF-alpha and IL-2 during T lymphocyte activation. This gene may also utilize a downstream in-frame translation start codon, and thus produce an isoform containing a shorter N-terminus. The shorter isoform has been shown to display weaker transforming activity. Alternate splicing results in multiple transcript variants that encode the same protein. It is required for lipopolysaccharide (LPS)-induced, TLR4-mediated activation of the MAPK/ERK pathway in macrophages, thus being critical for production of the proinflammatory cytokine TNF-alpha (TNF) during immune responses. Involved in the regulation of T-helper cell differentiation and IFNG expression in T-cells. Involved in mediating host resistance to bacterial infection through negative regulation of type I interferon (IFN) production. In vitro, activates MAPK/ERK pathway in response to IL1 in an IRAK1-independent manner, leading to up-regulation of IL8 and CCL4. Transduces CD40 and TNFRSF1A signals that activate ERK in B-cells and macrophages, and thus may play a role in the regulation of immunoglobulin production. May also play a role in the transduction of TNF signals that activate JNK and NF-kappa-B in some cell types. In adipocytes, activates MAPK/ERK pathway in an IKBKB-dependent manner in response to IL1B and TNF, but not insulin, leading to induction of lipolysis. Plays a role in the cell cycle. Isoform 1 shows some transforming activity, although it is much weaker than that of the activated oncogenic variant.
The expression profiles are the same for all samples: no differential expression, few reads, most of them mapping to exons at the 3' end.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 11 | 16 | 12 | 12 | 15 | 16 | 28 | 26 | 22 | |
Read_Max | R. aegyptiacus | Transcriptome | 7 | 13 | 8 | 9 | 13 | 16 | 3 | 18 | 12 | |
Read_Max | R. aegyptiacus | Genome | 26 | 12 | 18 | 19 | 14 | 12 | 11 | 28 | 28 | |
DESeq | H. sapiens | Genome | 167.43 | 235.57 | 203.54 | 188.07 | 187.98 | 281.29 | 255.39 | 271.98 | 272.84 | |
DESeq | R. aegyptiacus | Transcriptome | 14.44 | 25.51 | 25.43 | 17.48 | 26.19 | 30.51 | 6.56 | 18.35 | 24.62 | |
DESeq | R. aegyptiacus | Genome | 128.91 | 101.31 | 158.44 | 150.1 | 136.17 | 182.76 | 112.45 | 124.49 | 180.66 |