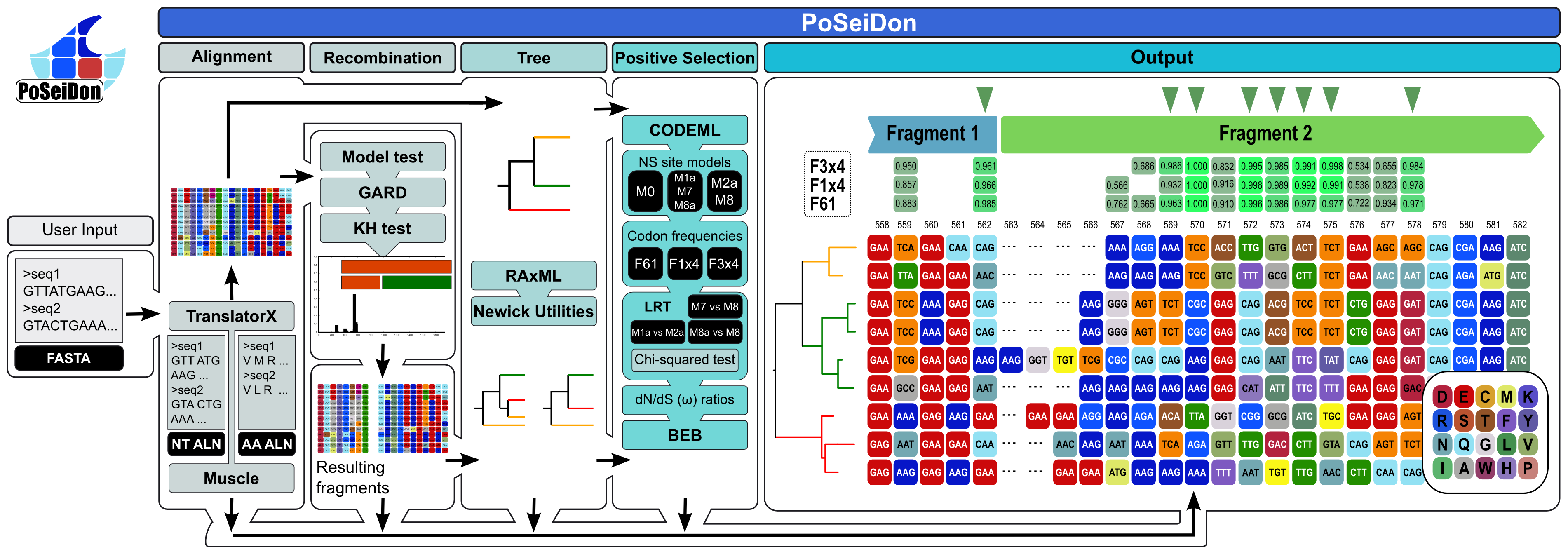
PoSeiDon (v1.1) parameter settings and executed commands
Commands
- Download commands full_aln
- Download commands fragment_1
- Download commands fragment_2
- Download commands fragment_3
Get configuration files used to run CodeML
Commands full_aln
################################# START PoSeiDon: 1488360010###############################PROJECT_DIR=/your/project/dir#CALCULATIONS ON FULL ALIGNMENT...#BUILD ALIGNMENT...translatorx.pl -i $PROJECT_DIR/bats_mx1.fna -p M -o $PROJECT_DIR/aln/bats_mx1#BUILD PHYLOGENY...raxmlHPC-PTHREADS-SSE3 -T 45 -f a -# 100 -x 1234 -p 1234 -s $PROJECT_DIR/aln/bats_mx1.nt_ali.nogaps.fasta -n nt -m GTRGAMMA -N 1000 -w $PROJECT_DIR/phylo/nw_display -v 50 -i 'font-size:11' -l 'font-size:16;font-family:helvetica;font-style:italic' -d 'stroke:black;fill:none;stroke-width:2;stroke-linecap:round' -Il -w 650 -b 'opacity:0' -s $PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.nt.corrected > $PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.nt.scale.svgnw_reroot $PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.nt.corrected > $PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.nt.corrected.rootedraxmlHPC-PTHREADS-SSE3 -T 45 -f a -# 100 -x 1234 -p 1234 -s $PROJECT_DIR/aln/bats_mx1.nt_ali.nogaps.fasta -n aa -m PROTGAMMAWAG -N 1000 -w $PROJECT_DIR/phylo/nw_display -v 50 -i 'font-size:11' -l 'font-size:16;font-family:helvetica;font-style:italic' -d 'stroke:black;fill:none;stroke-width:2;stroke-linecap:round' -Il -w 650 -b 'opacity:0' -s $PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.aa.corrected > $PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.aa.scale.svgnw_reroot $PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.aa.corrected > $PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.aa.corrected.rooted#RUN MODELSELECTION...(echo "$PROJECT_DIR/aln/bats_mx1.nt_ali.nogaps.fasta"; echo "$PROJECT_DIR/phylo/RAxML_bipartitionsBranchLabels.nt.unshod.tree"; echo "4"; echo "1"; echo "0.05"; echo "$PROJECT_DIR/model_selection/bats_mx1.nt_ali.nogaps.fasta.result") | mpirun -np 45 HYPHYMPI ModelTest.bf#RUN GARD...(echo "$PROJECT_DIR/aln/bats_mx1.nt_ali.nogaps.fasta"; echo "$PROJECT_DIR/aln/bats_mx1.nt_ali.nogaps.fasta"; echo "010010"; echo "2"; echo "3"; echo "$PROJECT_DIR/gard/gard") | mpirun -np 45 HYPHYMPI GARD.bf#GARDProcessor(echo "$PROJECT_DIR/gard/gard_finalout"; echo "$PROJECT_DIR/gard/gard_splits") | mpirun -np 45 HYPHYMPI GARDProcessor.bf#RUN CODEML ON FULL ALIGNMENT...codeml $PROJECT_DIR/codeml/F61/M0/codeml.ctlcodeml $PROJECT_DIR/codeml/F61/M1a/codeml.ctlcodeml $PROJECT_DIR/codeml/F61/M2a/codeml.ctlcodeml $PROJECT_DIR/codeml/F61/M7/codeml.ctlcodeml $PROJECT_DIR/codeml/F61/M8/codeml.ctlcodeml $PROJECT_DIR/codeml/F61/M8a/codeml.ctlcodeml $PROJECT_DIR/codeml/F1X4/M0/codeml.ctlcodeml $PROJECT_DIR/codeml/F1X4/M1a/codeml.ctlcodeml $PROJECT_DIR/codeml/F1X4/M2a/codeml.ctlcodeml $PROJECT_DIR/codeml/F1X4/M7/codeml.ctlcodeml $PROJECT_DIR/codeml/F1X4/M8/codeml.ctlcodeml $PROJECT_DIR/codeml/F1X4/M8a/codeml.ctlcodeml $PROJECT_DIR/codeml/F3X4/M0/codeml.ctlcodeml $PROJECT_DIR/codeml/F3X4/M1a/codeml.ctlcodeml $PROJECT_DIR/codeml/F3X4/M2a/codeml.ctlcodeml $PROJECT_DIR/codeml/F3X4/M7/codeml.ctlcodeml $PROJECT_DIR/codeml/F3X4/M8/codeml.ctlcodeml $PROJECT_DIR/codeml/F3X4/M8a/codeml.ctl#BREAKPOINTS DETECTED, ALIGNMENT ACCORDINGLY SPLITTEDCommands fragment_1
#BREAKPOINTS DETECTED, ALIGNMENT ACCORDINGLY SPLITTED#################################################CALCULATIONS ON FRAGMENT 1#BUILD PHYLOGENY fragment_1raxmlHPC-PTHREADS-SSE3 -T 45 -f a -# 100 -x 1234 -p 1234 -s $PROJECT_DIR/fragments/fragment_1/aln/bats_mx1.nt_ali.nogaps.fasta -n nt -m GTRGAMMA -N 1000 -w $PROJECT_DIR/fragments/fragment_1/phylo/nw_display -v 50 -i 'font-size:11' -l 'font-size:16;font-family:helvetica;font-style:italic' -d 'stroke:black;fill:none;stroke-width:2;stroke-linecap:round' -Il -w 650 -b 'opacity:0' -s $PROJECT_DIR/fragments/fragment_1/phylo/RAxML_bipartitionsBranchLabels.nt.corrected > $PROJECT_DIR/fragments/fragment_1/phylo/RAxML_bipartitionsBranchLabels.nt.scale.svgnw_reroot $PROJECT_DIR/fragments/fragment_1/phylo/RAxML_bipartitionsBranchLabels.nt.corrected > $PROJECT_DIR/fragments/fragment_1/phylo/RAxML_bipartitionsBranchLabels.nt.corrected.rootedraxmlHPC-PTHREADS-SSE3 -T 45 -f a -# 100 -x 1234 -p 1234 -s $PROJECT_DIR/fragments/fragment_1/aln/bats_mx1.aa_ali.nogaps.fasta -n aa -m PROTGAMMAWAG -N 1000 -w $PROJECT_DIR/fragments/fragment_1/phylo/nw_display -v 50 -i 'font-size:11' -l 'font-size:16;font-family:helvetica;font-style:italic' -d 'stroke:black;fill:none;stroke-width:2;stroke-linecap:round' -Il -w 650 -b 'opacity:0' -s $PROJECT_DIR/fragments/fragment_1/phylo/RAxML_bipartitionsBranchLabels.aa.corrected > $PROJECT_DIR/fragments/fragment_1/phylo/RAxML_bipartitionsBranchLabels.aa.scale.svgnw_reroot $PROJECT_DIR/fragments/fragment_1/phylo/RAxML_bipartitionsBranchLabels.aa.corrected > $PROJECT_DIR/fragments/fragment_1/phylo/RAxML_bipartitionsBranchLabels.aa.corrected.rooted#RUN CODEML ON fragment_1codeml $PROJECT_DIR/fragments/fragment_1/codeml/F61/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F61/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F61/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F61/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F61/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F61/M8a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F1X4/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F1X4/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F1X4/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F1X4/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F1X4/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F1X4/M8a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F3X4/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F3X4/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F3X4/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F3X4/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F3X4/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_1/codeml/F3X4/M8a/codeml.ctlCommands fragment_2
#BREAKPOINTS DETECTED, ALIGNMENT ACCORDINGLY SPLITTED#################################################CALCULATIONS ON FRAGMENT 2#BUILD PHYLOGENY fragment_2raxmlHPC-PTHREADS-SSE3 -T 45 -f a -# 100 -x 1234 -p 1234 -s $PROJECT_DIR/fragments/fragment_2/aln/bats_mx1.nt_ali.nogaps.fasta -n nt -m GTRGAMMA -N 1000 -w $PROJECT_DIR/fragments/fragment_2/phylo/nw_display -v 50 -i 'font-size:11' -l 'font-size:16;font-family:helvetica;font-style:italic' -d 'stroke:black;fill:none;stroke-width:2;stroke-linecap:round' -Il -w 650 -b 'opacity:0' -s $PROJECT_DIR/fragments/fragment_2/phylo/RAxML_bipartitionsBranchLabels.nt.corrected > $PROJECT_DIR/fragments/fragment_2/phylo/RAxML_bipartitionsBranchLabels.nt.scale.svgnw_reroot $PROJECT_DIR/fragments/fragment_2/phylo/RAxML_bipartitionsBranchLabels.nt.corrected > $PROJECT_DIR/fragments/fragment_2/phylo/RAxML_bipartitionsBranchLabels.nt.corrected.rootedraxmlHPC-PTHREADS-SSE3 -T 45 -f a -# 100 -x 1234 -p 1234 -s $PROJECT_DIR/fragments/fragment_2/aln/bats_mx1.aa_ali.nogaps.fasta -n aa -m PROTGAMMAWAG -N 1000 -w $PROJECT_DIR/fragments/fragment_2/phylo/nw_display -v 50 -i 'font-size:11' -l 'font-size:16;font-family:helvetica;font-style:italic' -d 'stroke:black;fill:none;stroke-width:2;stroke-linecap:round' -Il -w 650 -b 'opacity:0' -s $PROJECT_DIR/fragments/fragment_2/phylo/RAxML_bipartitionsBranchLabels.aa.corrected > $PROJECT_DIR/fragments/fragment_2/phylo/RAxML_bipartitionsBranchLabels.aa.scale.svgnw_reroot $PROJECT_DIR/fragments/fragment_2/phylo/RAxML_bipartitionsBranchLabels.aa.corrected > $PROJECT_DIR/fragments/fragment_2/phylo/RAxML_bipartitionsBranchLabels.aa.corrected.rooted#RUN CODEML ON fragment_2codeml $PROJECT_DIR/fragments/fragment_2/codeml/F61/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F61/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F61/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F61/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F61/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F61/M8a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F1X4/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F1X4/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F1X4/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F1X4/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F1X4/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F1X4/M8a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F3X4/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F3X4/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F3X4/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F3X4/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F3X4/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_2/codeml/F3X4/M8a/codeml.ctlCommands fragment_3
#BREAKPOINTS DETECTED, ALIGNMENT ACCORDINGLY SPLITTED#################################################CALCULATIONS ON FRAGMENT 3#BUILD PHYLOGENY fragment_3raxmlHPC-PTHREADS-SSE3 -T 45 -f a -# 100 -x 1234 -p 1234 -s $PROJECT_DIR/fragments/fragment_3/aln/bats_mx1.nt_ali.nogaps.fasta -n nt -m GTRGAMMA -N 1000 -w $PROJECT_DIR/fragments/fragment_3/phylo/nw_display -v 50 -i 'font-size:11' -l 'font-size:16;font-family:helvetica;font-style:italic' -d 'stroke:black;fill:none;stroke-width:2;stroke-linecap:round' -Il -w 650 -b 'opacity:0' -s $PROJECT_DIR/fragments/fragment_3/phylo/RAxML_bipartitionsBranchLabels.nt.corrected > $PROJECT_DIR/fragments/fragment_3/phylo/RAxML_bipartitionsBranchLabels.nt.scale.svgnw_reroot $PROJECT_DIR/fragments/fragment_3/phylo/RAxML_bipartitionsBranchLabels.nt.corrected > $PROJECT_DIR/fragments/fragment_3/phylo/RAxML_bipartitionsBranchLabels.nt.corrected.rootedraxmlHPC-PTHREADS-SSE3 -T 45 -f a -# 100 -x 1234 -p 1234 -s $PROJECT_DIR/fragments/fragment_3/aln/bats_mx1.aa_ali.nogaps.fasta -n aa -m PROTGAMMAWAG -N 1000 -w $PROJECT_DIR/fragments/fragment_3/phylo/nw_display -v 50 -i 'font-size:11' -l 'font-size:16;font-family:helvetica;font-style:italic' -d 'stroke:black;fill:none;stroke-width:2;stroke-linecap:round' -Il -w 650 -b 'opacity:0' -s $PROJECT_DIR/fragments/fragment_3/phylo/RAxML_bipartitionsBranchLabels.aa.corrected > $PROJECT_DIR/fragments/fragment_3/phylo/RAxML_bipartitionsBranchLabels.aa.scale.svgnw_reroot $PROJECT_DIR/fragments/fragment_3/phylo/RAxML_bipartitionsBranchLabels.aa.corrected > $PROJECT_DIR/fragments/fragment_3/phylo/RAxML_bipartitionsBranchLabels.aa.corrected.rooted#RUN CODEML ON fragment_3codeml $PROJECT_DIR/fragments/fragment_3/codeml/F61/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F61/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F61/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F61/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F61/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F61/M8a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F1X4/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F1X4/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F1X4/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F1X4/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F1X4/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F1X4/M8a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F3X4/M0/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F3X4/M1a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F3X4/M2a/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F3X4/M7/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F3X4/M8/codeml.ctlcodeml $PROJECT_DIR/fragments/fragment_3/codeml/F3X4/M8a/codeml.ctlTools executed by PoSeiDon
- TranslatorX (v1.1), Abascal et al. (2010); 20435676
- Muscle (v3.8.31), Edgar (2004); 15034147
- RAxML (v8.0.25), Stamatakis (2014); 24451623
- Newick Utilities (v1.6), Junier and Zdobnov (2010); 20472542
- MODELTEST , Posada and Crandall (1998); 9918953
- HyPhy (v2.2), Pond et al. (2005); 15509596
- GARD , Pond et al. (2006); 17110367
- PaML/CodeML (v4.8), Yang (2007); 17483113
- Ruby (v2.3.1)
- Inkscape (v0.48.5)
- pdfTeX (v3.14)
The pipeline