IFNAR2
| Name | Synonyms | Full Name | RefSeq ID | Description (.pdf) | IGV-img (humangenome) | Sashimi-img (humangenome) | UCSC-img (humangenome) | IGV-img (batgenome) |
|---|---|---|---|---|---|---|---|---|
| IFNAR2 | IFN-R-2 | interferon (alpha, beta and omega) receptor 2 |  |
 |
 |
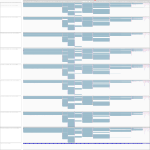 |
Download all snapshots for IGV, UCSC and Sashimi (zip archive)
Description
The protein encoded by this gene is a type I membrane protein that forms one of the two chains of a receptor for interferons alpha and beta. Binding and activation of the receptor stimulates Janus protein kinases, which in turn phosphorylate several proteins, including STAT1 and STAT2.
Marburg and Ebola virus infected human cells cells after 23 h show diverging directions of expression, albeit below 2 times fold change. In bat cells this gene is also expressed, but the reads could only be mapped by segemehl.
Maximum read counts and DESeq normalized read counts for human and bat cell lines
| Source | Species | Mapping on | Mock3h | Mock7h | Mock23h | EBOV3h | EBOV7h | EBOV23h | MARV3h | MARV7h | MARV23h | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
Read_Max | H. sapiens | Genome | 118 | 109 | 116 | 137 | 165 | 110 | 140 | 114 | 216 | |
Read_Max | R. aegyptiacus | Transcriptome | 290 | 280 | 291 | 289 | 233 | 288 | 254 | 375 | 345 | |
Read_Max | R. aegyptiacus | Genome | 341 | 305 | 262 | 274 | 208 | 174 | 226 | 464 | 397 | |
DESeq | H. sapiens | Genome | 1865.16 | 1362.53 | 1495.2 | 1662.98 | 1529.35 | 1118.22 | 1572.07 | 1440.63 | 2604.11 | |
DESeq | R. aegyptiacus | Transcriptome | 1563.66 | 1540.44 | 1949.35 | 1618.07 | 1756.43 | 1571.61 | 1833.13 | 1711.25 | 1816.02 | |
DESeq | R. aegyptiacus | Genome | 2249.88 | 2322.78 | 2638.81 | 2296.89 | 2480.05 | 2272.72 | 2671.89 | 2432.76 | 2568.58 |