Attention! PoSeiDon detected recombination in your alignment
Before you proceed, please be aware that recombination events can have a profound impact on the calculated tree topologies and the positive selection detection. Especially, all results based on the analyses of short fragments resulting from putative recombination events should be further investigated with suspicion.
Please be aware that you defined to use break points for further calculations even if the KH test showed no significant topological incongruence! ("Use insignificant break points")
Please use the navigation or click on the alignment links below to check out the whole positive selection analysis.
Recombination test with GARD
GARD found evidence of 2 breakpoints
Detailed recombination output
| KH-test
At p = 0.01 there are 0 significant breakpoints At p = 0.05 there are 0 significant breakpoints At p = 0.1 there are 0 significant breakpoints Mean splits identify: 0.2962962962962963 |
Resulting tree topologies
Trees on nucleotide level based on the full alignment and alignment fragments are shown.
Full alignment Go to alignment | Fragment 1 Go to alignment |
Fragment 2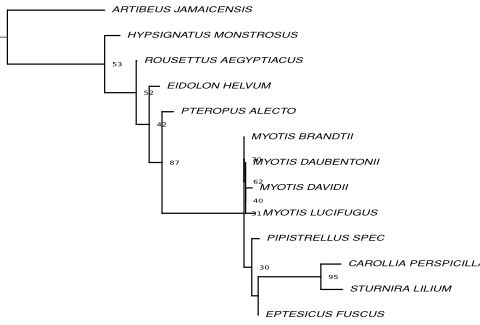 Go to alignment | Fragment 3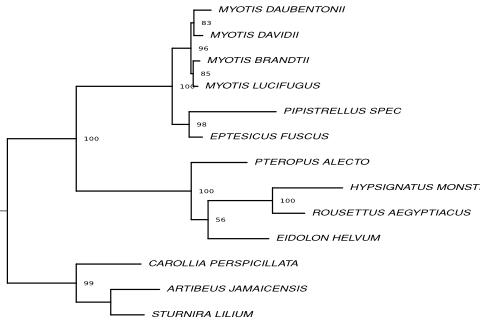 Go to alignment |
