Why DNA Methylation Matters – When we think about DNA, we often think of it as a static genomic sequence. But in reality, additional regulatory layers, known as epigenetic mechanisms, play a crucial role in controlling when and where genes are expressed. By adding small chemical groups, methyl groups are attached to specific DNA bases, predominantly at cytosine residues within CpG dinucleotides. This DNA methylation is essential for normal development, cellular differentiation, and the maintenance of stable gene expression patterns across organs.
DNA Methylation Changes in Cancer – In cancer, these tightly regulated methylation patterns are frequently disturbed. Tumour cells often exhibit aberrant DNA methylation. Typical cancer-specific changes include hypermethylation of the promoter regions of tumour suppressor genes, in combination with genome-wide hypomethylation. Such alterations can contribute directly to uncontrolled cell growth, genomic instability, and tumour progression.
Importantly, such methylation changes often appear very early during cancer development, sometimes even before the symptoms arise. This makes them particularly attractive as biomarkers for cancer detection, diagnosis, and disease monitoring.
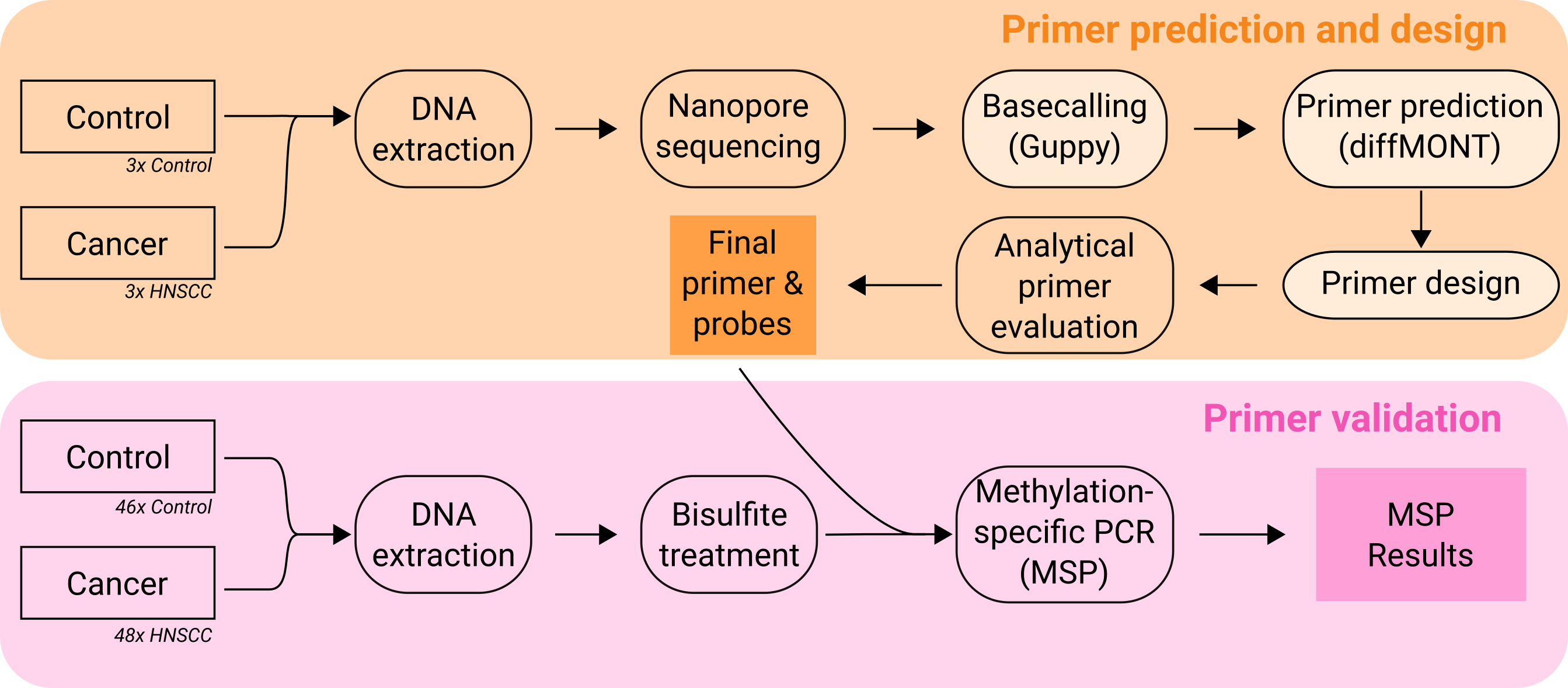
Turning Epigenetic Signals into Diagnostic Tests – Despite their promise for usage as biomarkers, translating methylation changes into robust diagnostic assays remains challenging. To tackle this challenge, we developed the Python-based tool diffMONT (https://github.com/rnajena/diffMONT), which predicts differentially methylated regions suitable for MSP analysis from nanopore sequencing data. Therewith, diffMONT combines modern nanopore DNA sequencing with the well established laboratory technique methylation-specific PCR (MSP).
Using a clinical dataset, which consisted of tissue samples from healthy individuals and patients with head and neck squamous cell carcinoma (HNSCC), we identified and validated methylation biomarkers for HNSCC. By leveraging genome-wide methylation information obtained from nanopore sequencing and converting it into targeted, cost-effective PCR assays, this approach provides a practical path from epigenetic discovery to clinically applicable diagnostics.
Contact: Daria Meyer (daria.meyer@uni-jena.de), Manja Marz (manja@uni-jena.de)
Original Publications:
Meyer, Daria; Barth, Emmanuel; Wiehle, Laura; Marz, Manja (2026) diffMONT: predicting methylation-specific PCR biomarkers based on nanopore sequencing data for clinical application. Bioinformatics (2026). doi: https://doi.org/10.1093/bioinformatics/btag039
Meyer, D., Hennig, A., Hums, AB. et al. Nanopore sequencing-derived methylation biomarker prediction for methylation-specific PCR in patients with head and neck squamous cell carcinoma. Clin Epigenet 17, 149 (2025). https://doi.org/10.1186/s13148-025-01960-7
Author: Daria Meyer
