The German Conference on Bioinformatics (GCB) is an “annual, international conference devoted to all areas of bioinformatics and meant as a platform for the whole bioinformatics community.” This year the conference took place at Bielefeld University with a variety of workshops and sessions on hot and fundamental research in bioinformatics. The session topics included data science, imaging, infrastructure & services, medical informatics, modeling, -omics, single cell research and structural biology.
This year the RNA Jena lab was honored by having four posters accepted for presentation in Bielefeld.
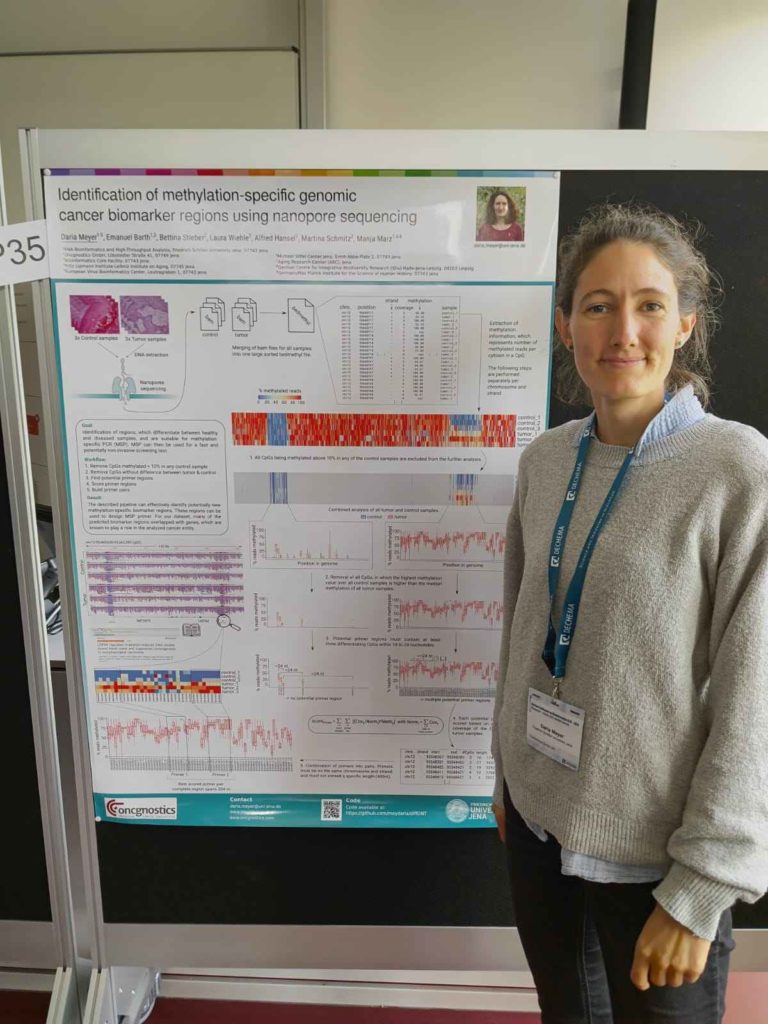
Photo of Daria Meyer presenting her poster titled “Identification of methylation-specific genomic cancer biomarker regions using nanopore sequencing.” Her algorithm (diffONT) identifies new methylation-specific biomarker regions by comparing paired healthy and cancerous Oxford Nanopore Technology (ONT) samples.
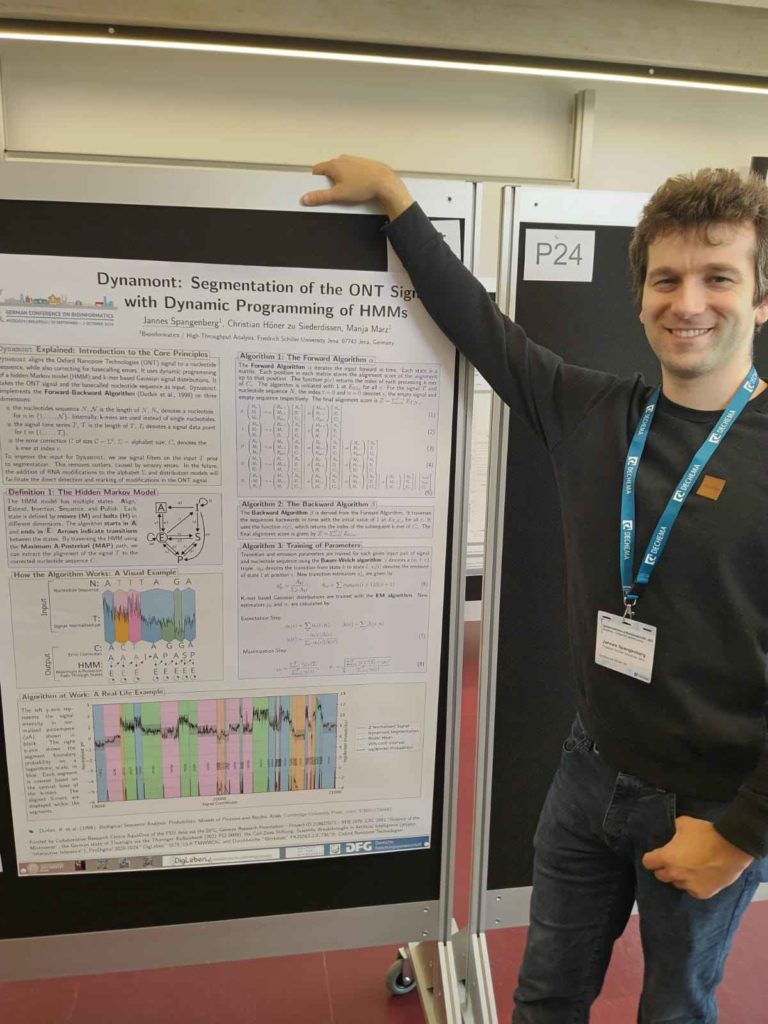
Photo of Jannes Spangenberg presenting his poster titled “Dynamont: Segmentation of the ONT Signal with Dynamic Programming of HMMs.” His recently developed resquiggle algorithm segments the ONT signals and corrects basecalling errors using dynamic programming of Hidden Markov Models and k-mer based Gaussian signal distributions. The use of more sophisticated models than are currently used will enable the segmentation and identification of RNA modifications within the ONT signal. Dynamont will be released soon — keep an eye on our GitHub or send us an email for more info!

Photo of Maria Schreiber presenting her poster titled “TOXIN-ANTITOXIN Systems: Seek and ye shall find.” She describes her new multi-step workflow to analyse the diversity of toxin-antitoxin systems within and between Bacterial and Archaeal species by leveraging homology searches and Rfam families. Her pilot project uses both dry lab and wet lab validation.

Photo of Shahram Saghaei presenting his poster “Building infrastructure for virus research” with an update on the progress of the VirJenDB, including integrating virus sequences and metadata from the European Nucleotide Archive (ENA) using the new NFDI resource Aruna. The database is funded by the DFG as a part of the NFDI4Microbiota to build FAIR and Open infrastructure for microbiome research.
Congrats to all presenters!
